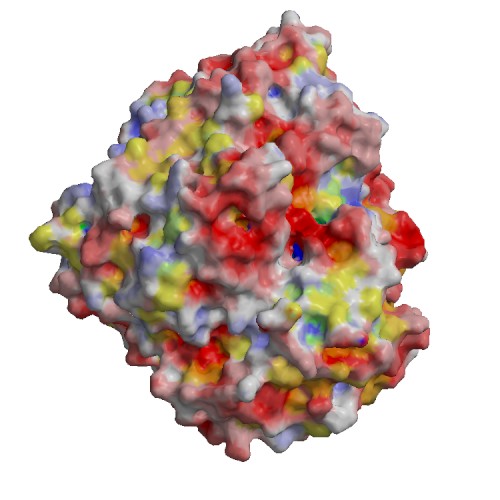
Functional site
| 1) | chain | B |
| residue | 108 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN B 1200 | |
| source | : AC2 | |
| 2) | chain | B |
| residue | 112 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN B 1200 | |
| source | : AC2 | |
| 3) | chain | B |
| residue | 189 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE ZN B 1200 | |
| source | : AC2 | |
| 4) | chain | B |
| residue | 205 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE DIO B 2001 | |
| source | : AC4 | |
| 5) | chain | B |
| residue | 477 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE DIO B 2001 | |
| source | : AC4 | |
| 6) | chain | B |
| residue | 479 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE DIO B 2001 | |
| source | : AC4 | |
| 7) | chain | B |
| residue | 111 | |
| type | ACT_SITE | |
| sequence | Q | |
| description | Proton acceptor => ECO:0000255|PROSITE-ProRule:PRU10096, ECO:0000305|PubMed:10684867, ECO:0000305|PubMed:17051221, ECO:0000305|PubMed:19321446, ECO:0000305|PubMed:23922390 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 8) | chain | B |
| residue | 108 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU10096, ECO:0000269|PubMed:17051221, ECO:0000269|PubMed:17613531, ECO:0000269|PubMed:18986166, ECO:0000269|PubMed:19321446, ECO:0007744|PDB:2G54, ECO:0007744|PDB:2JG4, ECO:0007744|PDB:3CWW, ECO:0007744|PDB:3N56, ECO:0007744|PDB:3N57 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 9) | chain | B |
| residue | 112 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU10096, ECO:0000269|PubMed:17051221, ECO:0000269|PubMed:17613531, ECO:0000269|PubMed:18986166, ECO:0000269|PubMed:19321446, ECO:0007744|PDB:2G54, ECO:0007744|PDB:2JG4, ECO:0007744|PDB:3CWW, ECO:0007744|PDB:3N56, ECO:0007744|PDB:3N57 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 10) | chain | B |
| residue | 189 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU10096, ECO:0000269|PubMed:17051221, ECO:0000269|PubMed:17613531, ECO:0000269|PubMed:18986166, ECO:0000269|PubMed:19321446, ECO:0007744|PDB:2G54, ECO:0007744|PDB:2JG4, ECO:0007744|PDB:3CWW, ECO:0007744|PDB:3N56, ECO:0007744|PDB:3N57 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 11) | chain | B |
| residue | 336 | |
| type | BINDING | |
| sequence | H | |
| description | in the exosite | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 12) | chain | B |
| residue | 359 | |
| type | BINDING | |
| sequence | L | |
| description | BINDING => ECO:0000269|PubMed:18986166 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 13) | chain | B |
| residue | 429 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000250|UniProtKB:P35559 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 14) | chain | B |
| residue | 895 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000250|UniProtKB:P35559 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 15) | chain | B |
| residue | 192 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine => ECO:0000250|UniProtKB:Q9JHR7 | |
| source | Swiss-Prot : SWS_FT_FI6 | |
| 16) | chain | B |
| residue | 697 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine => ECO:0000250|UniProtKB:Q9JHR7 | |
| source | Swiss-Prot : SWS_FT_FI6 | |