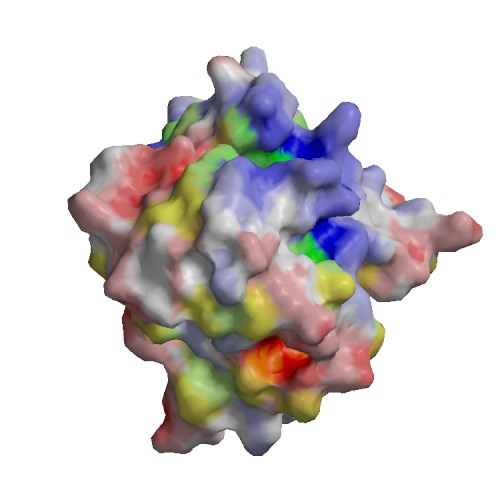
Functional site
| 1) | chain | B |
| residue | 7 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE DZF B 187 | |
| source | : AC2 | |
| 2) | chain | B |
| residue | 8 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE DZF B 187 | |
| source | : AC2 | |
| 3) | chain | B |
| residue | 30 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE DZF B 187 | |
| source | : AC2 | |
| 4) | chain | B |
| residue | 31 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE DZF B 187 | |
| source | : AC2 | |
| 5) | chain | B |
| residue | 34 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE DZF B 187 | |
| source | : AC2 | |
| 6) | chain | B |
| residue | 35 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE DZF B 187 | |
| source | : AC2 | |
| 7) | chain | B |
| residue | 64 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE DZF B 187 | |
| source | : AC2 | |
| 8) | chain | B |
| residue | 70 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE DZF B 187 | |
| source | : AC2 | |
| 9) | chain | B |
| residue | 115 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE DZF B 187 | |
| source | : AC2 | |
| 10) | chain | B |
| residue | 121 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE DZF B 187 | |
| source | : AC2 | |
| 11) | chain | B |
| residue | 136 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE DZF B 187 | |
| source | : AC2 | |
| 12) | chain | B |
| residue | 23 | |
| type | catalytic | |
| sequence | P | |
| description | 490 | |
| source | MCSA : MCSA2 | |
| 13) | chain | B |
| residue | 31 | |
| type | catalytic | |
| sequence | F | |
| description | 490 | |
| source | MCSA : MCSA2 | |
| 14) | chain | B |
| residue | 117 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000269|PubMed:15039552, ECO:0000269|PubMed:16222560, ECO:0000269|PubMed:19478082 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 15) | chain | B |
| residue | 10 | |
| type | BINDING | |
| sequence | V | |
| description | BINDING => ECO:0000269|PubMed:15039552, ECO:0000269|PubMed:16222560, ECO:0000269|PubMed:19478082 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 16) | chain | B |
| residue | 16 | |
| type | BINDING | |
| sequence | I | |
| description | BINDING => ECO:0000269|PubMed:15039552, ECO:0000269|PubMed:16222560, ECO:0000269|PubMed:19478082 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 17) | chain | B |
| residue | 55 | |
| type | BINDING | |
| sequence | K | |
| description | BINDING => ECO:0000269|PubMed:15039552, ECO:0000269|PubMed:16222560, ECO:0000269|PubMed:19478082 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 18) | chain | B |
| residue | 77 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000269|PubMed:15039552, ECO:0000269|PubMed:16222560, ECO:0000269|PubMed:19478082 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 19) | chain | B |
| residue | 31 | |
| type | BINDING | |
| sequence | F | |
| description | BINDING => ECO:0000305|PubMed:2248959 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 20) | chain | B |
| residue | 65 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000305|PubMed:2248959 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 21) | chain | B |
| residue | 71 | |
| type | BINDING | |
| sequence | I | |
| description | BINDING => ECO:0000305|PubMed:2248959 | |
| source | Swiss-Prot : SWS_FT_FI2 | |