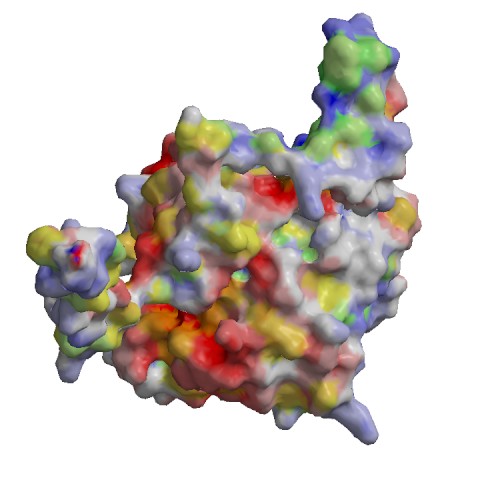
Functional site
| 1) | chain | D |
| residue | 160 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE ZN D1405 | |
| source | : AC4 | |
| 2) | chain | D |
| residue | 217 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN D1405 | |
| source | : AC4 | |
| 3) | chain | D |
| residue | 299 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN D1405 | |
| source | : AC4 | |
| 4) | chain | D |
| residue | 300 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN D1405 | |
| source | : AC4 | |
| 5) | chain | D |
| residue | 217 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000250|UniProtKB:Q93088, ECO:0000255|PROSITE-ProRule:PRU00333 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 6) | chain | D |
| residue | 299 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000250|UniProtKB:Q93088, ECO:0000255|PROSITE-ProRule:PRU00333 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 7) | chain | D |
| residue | 300 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000250|UniProtKB:Q93088, ECO:0000255|PROSITE-ProRule:PRU00333 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 8) | chain | D |
| residue | 40 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine => ECO:0000250|UniProtKB:O35490 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 9) | chain | D |
| residue | 93 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine => ECO:0000250|UniProtKB:O35490 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 10) | chain | D |
| residue | 98 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine => ECO:0000250|UniProtKB:O35490 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 11) | chain | D |
| residue | 232 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine => ECO:0000250|UniProtKB:O35490 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 12) | chain | D |
| residue | 241 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine => ECO:0000250|UniProtKB:O35490 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 13) | chain | D |
| residue | 340 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine => ECO:0000250|UniProtKB:O35490 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 14) | chain | D |
| residue | 377 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine => ECO:0000250|UniProtKB:O35490 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 15) | chain | D |
| residue | 330 | |
| type | MOD_RES | |
| sequence | S | |
| description | Phosphoserine => ECO:0007744|PubMed:22673903 | |
| source | Swiss-Prot : SWS_FT_FI3 | |