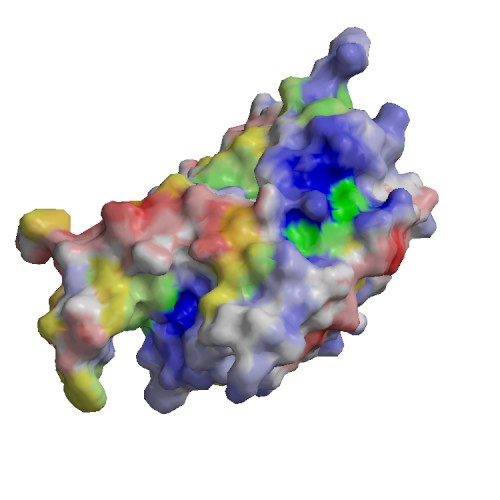
Functional site
| 1) | chain | A |
| residue | 121 | |
| type | ACT_SITE | |
| sequence | H | |
| description | ACT_SITE => ECO:0000250|UniProtKB:P29466 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 2) | chain | A |
| residue | 163 | |
| type | ACT_SITE | |
| sequence | C | |
| description | ACT_SITE => ECO:0000250|UniProtKB:P29466 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 3) | chain | A |
| residue | 163 | |
| type | MOD_RES | |
| sequence | C | |
| description | S-nitrosocysteine; in inhibited form => ECO:0000269|PubMed:10213689 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 4) | chain | A |
| residue | 62 | |
| type | catalytic | |
| sequence | T | |
| description | 815 | |
| source | MCSA : MCSA1 | |
| 5) | chain | A |
| residue | 63 | |
| type | catalytic | |
| sequence | S | |
| description | 815 | |
| source | MCSA : MCSA1 | |
| 6) | chain | A |
| residue | 121 | |
| type | catalytic | |
| sequence | H | |
| description | 815 | |
| source | MCSA : MCSA1 | |
| 7) | chain | A |
| residue | 122 | |
| type | catalytic | |
| sequence | G | |
| description | 815 | |
| source | MCSA : MCSA1 | |
| 8) | chain | A |
| residue | 163 | |
| type | catalytic | |
| sequence | C | |
| description | 815 | |
| source | MCSA : MCSA1 | |
| 9) | chain | A |
| residue | 207 | |
| type | MOD_RES | |
| sequence | R | |
| description | (Microbial infection) ADP-riboxanated arginine => ECO:0000269|PubMed:35338844, ECO:0000269|PubMed:35446120, ECO:0000269|PubMed:36423631 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 10) | chain | A |
| residue | 108-122 | |
| type | prosite | |
| sequence | HSKRSSFVCVLLSHG | |
| description | CASPASE_HIS Caspase family histidine active site. HskrsSfvCvLLSHG | |
| source | prosite : PS01121 | |
| 11) | chain | A |
| residue | 154-165 | |
| type | prosite | |
| sequence | KPKLFIIQACRG | |
| description | CASPASE_CYS Caspase family cysteine active site. KPKLFIIQACRG | |
| source | prosite : PS01122 | |