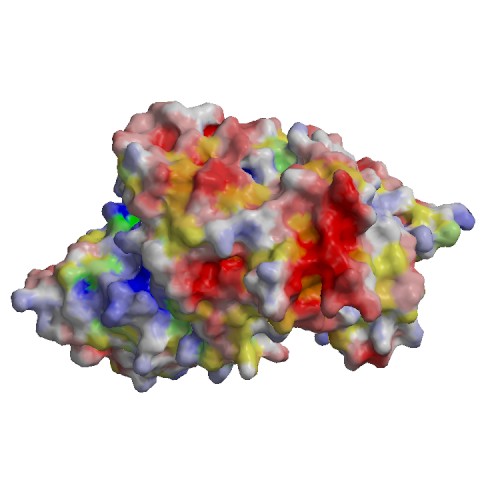
Functional site
| 1) | chain | A |
| residue | 53 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 2) | chain | A |
| residue | 54 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 58 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 4) | chain | A |
| residue | 59 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 5) | chain | A |
| residue | 60 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 6) | chain | A |
| residue | 79 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 7) | chain | A |
| residue | 82 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 8) | chain | A |
| residue | 83 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 9) | chain | A |
| residue | 85 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 10) | chain | A |
| residue | 162 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 11) | chain | A |
| residue | 333 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 12) | chain | A |
| residue | 369 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 13) | chain | A |
| residue | 433 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE HEM A 601 | |
| source | : AC1 | |
| 14) | chain | A |
| residue | 40 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 602 | |
| source | : AC2 | |
| 15) | chain | A |
| residue | 43 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE HEM A 602 | |
| source | : AC2 | |
| 16) | chain | A |
| residue | 46 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE HEM A 602 | |
| source | : AC2 | |
| 17) | chain | A |
| residue | 55 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE HEM A 602 | |
| source | : AC2 | |
| 18) | chain | A |
| residue | 56 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 602 | |
| source | : AC2 | |
| 19) | chain | A |
| residue | 65 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEM A 602 | |
| source | : AC2 | |
| 20) | chain | A |
| residue | 68 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEM A 602 | |
| source | : AC2 | |
| 21) | chain | A |
| residue | 69 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 602 | |
| source | : AC2 | |
| 22) | chain | A |
| residue | 88 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE HEM A 602 | |
| source | : AC2 | |
| 23) | chain | A |
| residue | 90 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE HEM A 602 | |
| source | : AC2 | |
| 24) | chain | A |
| residue | 5 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE HEM A 603 | |
| source | : AC3 | |
| 25) | chain | A |
| residue | 9 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 603 | |
| source | : AC3 | |
| 26) | chain | A |
| residue | 17 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE HEM A 603 | |
| source | : AC3 | |
| 27) | chain | A |
| residue | 35 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE HEM A 603 | |
| source | : AC3 | |
| 28) | chain | A |
| residue | 36 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEM A 603 | |
| source | : AC3 | |
| 29) | chain | A |
| residue | 39 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEM A 603 | |
| source | : AC3 | |
| 30) | chain | A |
| residue | 40 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 603 | |
| source | : AC3 | |
| 31) | chain | A |
| residue | 69 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 603 | |
| source | : AC3 | |
| 32) | chain | A |
| residue | 91 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE HEM A 603 | |
| source | : AC3 | |
| 33) | chain | A |
| residue | 227 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE HEM A 603 | |
| source | : AC3 | |
| 34) | chain | A |
| residue | 6 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 35) | chain | A |
| residue | 9 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 36) | chain | A |
| residue | 15 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 37) | chain | A |
| residue | 18 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 38) | chain | A |
| residue | 19 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 39) | chain | A |
| residue | 29 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 40) | chain | A |
| residue | 66 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 41) | chain | A |
| residue | 70 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 42) | chain | A |
| residue | 71 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 43) | chain | A |
| residue | 72 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 44) | chain | A |
| residue | 293 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 45) | chain | A |
| residue | 467 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 46) | chain | A |
| residue | 468 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 47) | chain | A |
| residue | 469 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 48) | chain | A |
| residue | 470 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM A 604 | |
| source | : AC4 | |
| 49) | chain | A |
| residue | 127 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 50) | chain | A |
| residue | 128 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 51) | chain | A |
| residue | 130 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 52) | chain | A |
| residue | 131 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 53) | chain | A |
| residue | 132 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 54) | chain | A |
| residue | 151 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 55) | chain | A |
| residue | 152 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 56) | chain | A |
| residue | 157 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 57) | chain | A |
| residue | 158 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 58) | chain | A |
| residue | 159 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 59) | chain | A |
| residue | 160 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 60) | chain | A |
| residue | 163 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 61) | chain | A |
| residue | 164 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 62) | chain | A |
| residue | 165 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 63) | chain | A |
| residue | 166 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 64) | chain | A |
| residue | 271 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 65) | chain | A |
| residue | 272 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 66) | chain | A |
| residue | 273 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 67) | chain | A |
| residue | 307 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 68) | chain | A |
| residue | 308 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 69) | chain | A |
| residue | 309 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 70) | chain | A |
| residue | 331 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 71) | chain | A |
| residue | 332 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 72) | chain | A |
| residue | 335 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 73) | chain | A |
| residue | 339 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 74) | chain | A |
| residue | 499 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 75) | chain | A |
| residue | 500 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 76) | chain | A |
| residue | 528 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 77) | chain | A |
| residue | 529 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 78) | chain | A |
| residue | 539 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 79) | chain | A |
| residue | 542 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 80) | chain | A |
| residue | 543 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 81) | chain | A |
| residue | 544 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 82) | chain | A |
| residue | 545 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 83) | chain | A |
| residue | 548 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE FAD A 605 | |
| source | : AC5 | |
| 84) | chain | A |
| residue | 397 | |
| type | ACT_SITE | |
| sequence | R | |
| description | Proton donor => ECO:0000250|UniProtKB:P0C278 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 85) | chain | A |
| residue | 9 | |
| type | BINDING | |
| sequence | H | |
| description | axial binding residue => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 86) | chain | A |
| residue | 19 | |
| type | BINDING | |
| sequence | H | |
| description | axial binding residue => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 87) | chain | A |
| residue | 40 | |
| type | BINDING | |
| sequence | H | |
| description | axial binding residue => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 88) | chain | A |
| residue | 56 | |
| type | BINDING | |
| sequence | H | |
| description | axial binding residue => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 89) | chain | A |
| residue | 59 | |
| type | BINDING | |
| sequence | H | |
| description | axial binding residue => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 90) | chain | A |
| residue | 69 | |
| type | BINDING | |
| sequence | H | |
| description | axial binding residue => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 91) | chain | A |
| residue | 72 | |
| type | BINDING | |
| sequence | H | |
| description | axial binding residue => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 92) | chain | A |
| residue | 83 | |
| type | BINDING | |
| sequence | H | |
| description | axial binding residue => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 93) | chain | A |
| residue | 15 | |
| type | BINDING | |
| sequence | C | |
| description | covalent => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 94) | chain | A |
| residue | 18 | |
| type | BINDING | |
| sequence | C | |
| description | covalent => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 95) | chain | A |
| residue | 36 | |
| type | BINDING | |
| sequence | C | |
| description | covalent => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 96) | chain | A |
| residue | 39 | |
| type | BINDING | |
| sequence | C | |
| description | covalent => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 97) | chain | A |
| residue | 65 | |
| type | BINDING | |
| sequence | C | |
| description | covalent => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 98) | chain | A |
| residue | 68 | |
| type | BINDING | |
| sequence | C | |
| description | covalent => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 99) | chain | A |
| residue | 79 | |
| type | BINDING | |
| sequence | C | |
| description | covalent => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 100) | chain | A |
| residue | 82 | |
| type | BINDING | |
| sequence | C | |
| description | covalent => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 101) | chain | A |
| residue | 71 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 102) | chain | A |
| residue | 165 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 103) | chain | A |
| residue | 196 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 104) | chain | A |
| residue | 360 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 105) | chain | A |
| residue | 372 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 106) | chain | A |
| residue | 373 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 107) | chain | A |
| residue | 499 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 108) | chain | A |
| residue | 539 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 109) | chain | A |
| residue | 542 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000250|UniProtKB:P83223 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 110) | chain | A |
| residue | 132 | |
| type | BINDING | |
| sequence | A | |
| description | BINDING => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 111) | chain | A |
| residue | 545 | |
| type | BINDING | |
| sequence | I | |
| description | BINDING => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 112) | chain | A |
| residue | 151 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 113) | chain | A |
| residue | 159 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 114) | chain | A |
| residue | 160 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 115) | chain | A |
| residue | 166 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 116) | chain | A |
| residue | 273 | |
| type | BINDING | |
| sequence | V | |
| description | BINDING => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 117) | chain | A |
| residue | 339 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 118) | chain | A |
| residue | 529 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 119) | chain | A |
| residue | 544 | |
| type | BINDING | |
| sequence | A | |
| description | BINDING => ECO:0000269|PubMed:10581549, ECO:0007744|PDB:1QO8 | |
| source | Swiss-Prot : SWS_FT_FI5 | |