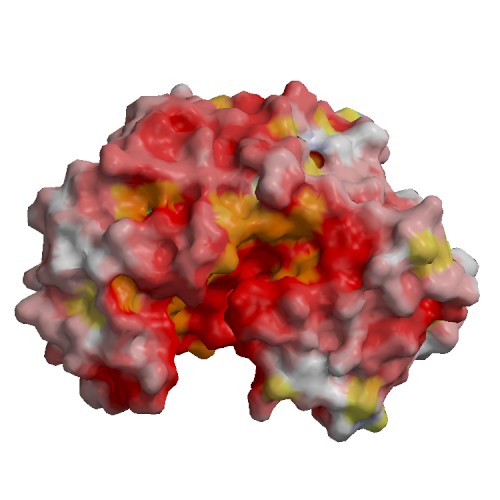
Functional site
| 1) | chain | E |
| residue | 30-41 | |
| type | prosite | |
| sequence | LNFDTGSADLWV | |
| description | ASP_PROTEASE Eukaryotic and viral aspartyl proteases active site. LNFDTGSADLWV | |
| source | prosite : PS00141 | |
| 2) | chain | E |
| residue | 210-221 | |
| type | prosite | |
| sequence | GIADTGTTLLLL | |
| description | ASP_PROTEASE Eukaryotic and viral aspartyl proteases active site. LNFDTGSADLWV | |
| source | prosite : PS00141 | |
| 3) | chain | E |
| residue | 33 | |
| type | ACT_SITE | |
| sequence | D | |
| description | ACT_SITE => ECO:0000255|PROSITE-ProRule:PRU01103, ECO:0000269|PubMed:5475460 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 4) | chain | E |
| residue | 213 | |
| type | ACT_SITE | |
| sequence | D | |
| description | ACT_SITE => ECO:0000255|PROSITE-ProRule:PRU01103, ECO:0000269|PubMed:5475460 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 5) | chain | E |
| residue | 3 | |
| type | CARBOHYD | |
| sequence | S | |
| description | O-linked (Man...) serine => ECO:0000269|PubMed:9836576, ECO:0000269|DOI:10.1021/ja973714r | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 6) | chain | E |
| residue | 7 | |
| type | CARBOHYD | |
| sequence | T | |
| description | O-linked (Man...) threonine => ECO:0000269|PubMed:9836576, ECO:0000269|DOI:10.1021/ja973714r | |
| source | Swiss-Prot : SWS_FT_FI3 | |