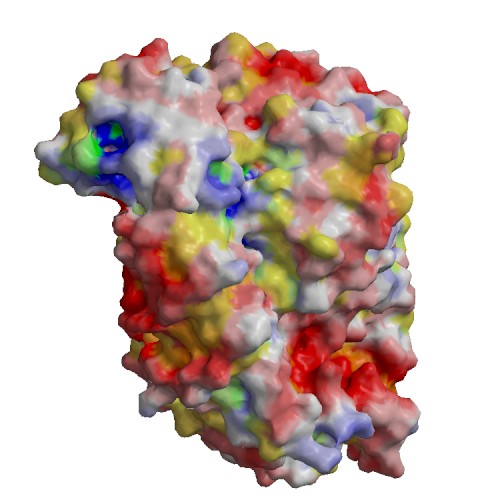
Functional site
| 1) | chain | B |
| residue | 297 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE ZN B 500 | |
| source | : AC1 | |
| 2) | chain | B |
| residue | 299 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN B 500 | |
| source | : AC1 | |
| 3) | chain | B |
| residue | 362 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN B 500 | |
| source | : AC1 | |
| 4) | chain | A |
| residue | 164 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 5) | chain | A |
| residue | 166 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 6) | chain | A |
| residue | 200 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 7) | chain | B |
| residue | 102 | |
| type | ||
| sequence | W | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 8) | chain | B |
| residue | 205 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 9) | chain | B |
| residue | 248 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 10) | chain | B |
| residue | 250 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 11) | chain | B |
| residue | 291 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 12) | chain | B |
| residue | 294 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 13) | chain | B |
| residue | 300 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 14) | chain | B |
| residue | 303 | |
| type | ||
| sequence | W | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 15) | chain | B |
| residue | 361 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE HFP B 501 | |
| source | : AC2 | |
| 16) | chain | A |
| residue | 166 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE 2C5 B 10 | |
| source | : AC3 | |
| 17) | chain | B |
| residue | 93 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE 2C5 B 10 | |
| source | : AC3 | |
| 18) | chain | B |
| residue | 96 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE 2C5 B 10 | |
| source | : AC3 | |
| 19) | chain | B |
| residue | 106 | |
| type | ||
| sequence | W | |
| description | BINDING SITE FOR RESIDUE 2C5 B 10 | |
| source | : AC3 | |
| 20) | chain | B |
| residue | 202 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE 2C5 B 10 | |
| source | : AC3 | |
| 21) | chain | B |
| residue | 297 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE 2C5 B 10 | |
| source | : AC3 | |
| 22) | chain | B |
| residue | 299 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE 2C5 B 10 | |
| source | : AC3 | |
| 23) | chain | B |
| residue | 359 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE 2C5 B 10 | |
| source | : AC3 | |
| 24) | chain | B |
| residue | 360 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE 2C5 B 10 | |
| source | : AC3 | |
| 25) | chain | B |
| residue | 361 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE 2C5 B 10 | |
| source | : AC3 | |
| 26) | chain | B |
| residue | 248 | |
| type | catalytic | |
| sequence | H | |
| description | 484 | |
| source | MCSA : MCSA1 | |
| 27) | chain | B |
| residue | 291 | |
| type | catalytic | |
| sequence | R | |
| description | 484 | |
| source | MCSA : MCSA1 | |
| 28) | chain | B |
| residue | 294 | |
| type | catalytic | |
| sequence | K | |
| description | 484 | |
| source | MCSA : MCSA1 | |
| 29) | chain | B |
| residue | 297 | |
| type | catalytic | |
| sequence | D | |
| description | 484 | |
| source | MCSA : MCSA1 | |
| 30) | chain | B |
| residue | 299 | |
| type | catalytic | |
| sequence | C | |
| description | 484 | |
| source | MCSA : MCSA1 | |
| 31) | chain | B |
| residue | 300 | |
| type | catalytic | |
| sequence | Y | |
| description | 484 | |
| source | MCSA : MCSA1 | |
| 32) | chain | B |
| residue | 352 | |
| type | catalytic | |
| sequence | D | |
| description | 484 | |
| source | MCSA : MCSA1 | |
| 33) | chain | B |
| residue | 359 | |
| type | catalytic | |
| sequence | D | |
| description | 484 | |
| source | MCSA : MCSA1 | |
| 34) | chain | B |
| residue | 362 | |
| type | catalytic | |
| sequence | H | |
| description | 484 | |
| source | MCSA : MCSA1 | |
| 35) | chain | B |
| residue | 248 | |
| type | BINDING | |
| sequence | H | |
| description | ||
| source | Swiss-Prot : SWS_FT_FI1 | |
| 36) | chain | B |
| residue | 291 | |
| type | BINDING | |
| sequence | R | |
| description | ||
| source | Swiss-Prot : SWS_FT_FI1 | |
| 37) | chain | B |
| residue | 300 | |
| type | BINDING | |
| sequence | Y | |
| description | ||
| source | Swiss-Prot : SWS_FT_FI1 | |
| 38) | chain | B |
| residue | 102 | |
| type | SITE | |
| sequence | W | |
| description | Important for selectivity against geranylgeranyl diphosphate => ECO:0000250|UniProtKB:P49356 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 39) | chain | B |
| residue | 297 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:18844669, ECO:0000269|PubMed:20056542, ECO:0000269|PubMed:9065406 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 40) | chain | B |
| residue | 299 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:18844669, ECO:0000269|PubMed:20056542, ECO:0000269|PubMed:9065406 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 41) | chain | B |
| residue | 362 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000269|PubMed:18844669, ECO:0000269|PubMed:20056542, ECO:0000269|PubMed:9065406 | |
| source | Swiss-Prot : SWS_FT_FI2 | |