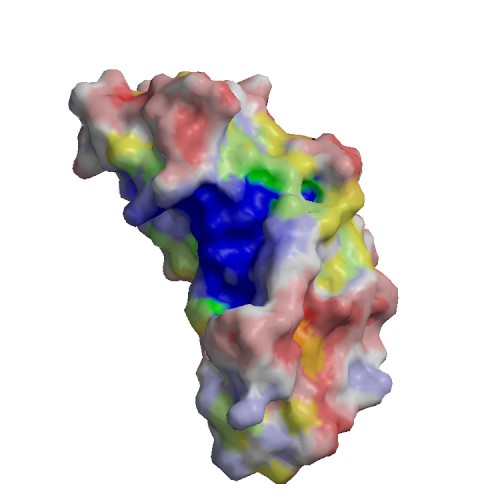
Functional site
| 1) | chain | A |
| residue | 12 | |
| type | ||
| sequence | X | |
| description | BINDING SITE FOR RESIDUE SO4 A 301 | |
| source | : AC1 | |
| 2) | chain | A |
| residue | 13 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE SO4 A 301 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 14 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE SO4 A 301 | |
| source | : AC1 | |
| 4) | chain | A |
| residue | 62 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE SO4 A 301 | |
| source | : AC1 | |
| 5) | chain | A |
| residue | 67 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE SO4 A 301 | |
| source | : AC1 | |
| 6) | chain | A |
| residue | 107 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 A 302 | |
| source | : AC2 | |
| 7) | chain | A |
| residue | 14 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE SO4 A 303 | |
| source | : AC3 | |
| 8) | chain | A |
| residue | 17 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE SO4 A 303 | |
| source | : AC3 | |
| 9) | chain | A |
| residue | 108 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE SO4 A 303 | |
| source | : AC3 | |
| 10) | chain | A |
| residue | 109 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE SO4 A 303 | |
| source | : AC3 | |
| 11) | chain | A |
| residue | 126 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE SO4 A 303 | |
| source | : AC3 | |
| 12) | chain | A |
| residue | 12 | |
| type | ||
| sequence | X | |
| description | BINDING SITE FOR RESIDUE SO3 A 401 | |
| source | : AC4 | |
| 13) | chain | A |
| residue | 60 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO3 A 401 | |
| source | : AC4 | |
| 14) | chain | A |
| residue | 94 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO3 A 401 | |
| source | : AC4 | |
| 15) | chain | A |
| residue | 113 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE CS A 501 | |
| source | : AC5 | |
| 16) | chain | A |
| residue | 113 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE CS A 501 | |
| source | : AC5 | |
| 17) | chain | A |
| residue | 114 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CS A 501 | |
| source | : AC5 | |
| 18) | chain | A |
| residue | 114 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CS A 501 | |
| source | : AC5 | |
| 19) | chain | A |
| residue | 116 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE CS A 501 | |
| source | : AC5 | |
| 20) | chain | A |
| residue | 116 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE CS A 501 | |
| source | : AC5 | |
| 21) | chain | A |
| residue | 67 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE CS A 502 | |
| source | : AC6 | |
| 22) | chain | A |
| residue | 38 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE CS A 503 | |
| source | : AC7 | |
| 23) | chain | A |
| residue | 43 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE CS A 503 | |
| source | : AC7 | |
| 24) | chain | A |
| residue | 26 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE CS A 504 | |
| source | : AC8 | |
| 25) | chain | A |
| residue | 64 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE CS A 505 | |
| source | : AC9 | |
| 26) | chain | A |
| residue | 87 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE CS A 505 | |
| source | : AC9 | |
| 27) | chain | A |
| residue | 12 | |
| type | ACT_SITE | |
| sequence | X | |
| description | Nucleophile; cysteine thioarsenate intermediate => ECO:0000255|PROSITE-ProRule:PRU01282, ECO:0000269|PubMed:11709171, ECO:0000269|PubMed:15295115, ECO:0000269|PubMed:7577935 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 28) | chain | A |
| residue | 8 | |
| type | SITE | |
| sequence | H | |
| description | Important for activity. Lowers pKa of the active site Cys => ECO:0000269|PubMed:8969183 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 29) | chain | A |
| residue | 60 | |
| type | SITE | |
| sequence | R | |
| description | Important for activity. Involved in arsenate binding and transition-state stabilization => ECO:0000269|PubMed:11709171, ECO:0000269|PubMed:14592722, ECO:0000269|PubMed:15295115 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 30) | chain | A |
| residue | 94 | |
| type | SITE | |
| sequence | R | |
| description | Important for activity. Involved in arsenate binding and transition-state stabilization => ECO:0000269|PubMed:11709171, ECO:0000269|PubMed:14592722, ECO:0000269|PubMed:15295115 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 31) | chain | A |
| residue | 107 | |
| type | SITE | |
| sequence | R | |
| description | Important for activity. Involved in arsenate binding and transition-state stabilization => ECO:0000269|PubMed:11709171, ECO:0000269|PubMed:14592722, ECO:0000269|PubMed:15295115 | |
| source | Swiss-Prot : SWS_FT_FI3 | |