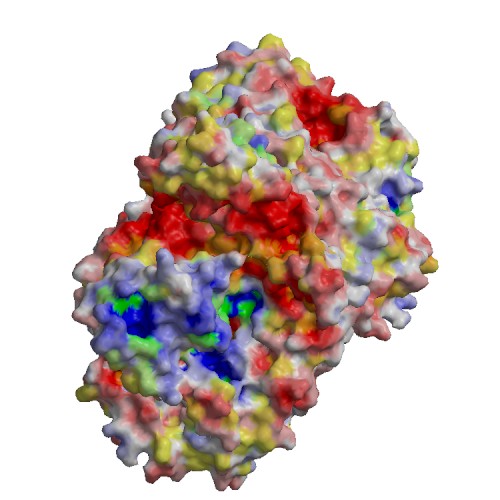
Functional site
| 1) | chain | A |
| residue | 24 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE MG A 251 | |
| source | : AC1 | |
| 2) | chain | A |
| residue | 40 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE MG A 251 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 42 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE MG A 251 | |
| source | : AC1 | |
| 4) | chain | C |
| residue | 24 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE MG C 253 | |
| source | : AC2 | |
| 5) | chain | C |
| residue | 40 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE MG C 253 | |
| source | : AC2 | |
| 6) | chain | C |
| residue | 42 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE MG C 253 | |
| source | : AC2 | |
| 7) | chain | A |
| residue | 19 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 8) | chain | A |
| residue | 20 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 9) | chain | A |
| residue | 21 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 10) | chain | A |
| residue | 22 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 11) | chain | A |
| residue | 23 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 12) | chain | A |
| residue | 24 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 13) | chain | A |
| residue | 25 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 14) | chain | A |
| residue | 35 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 15) | chain | A |
| residue | 36 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 16) | chain | A |
| residue | 37 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 17) | chain | A |
| residue | 38 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 18) | chain | A |
| residue | 39 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 19) | chain | A |
| residue | 41 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 20) | chain | A |
| residue | 42 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 21) | chain | A |
| residue | 68 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 22) | chain | A |
| residue | 69 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 23) | chain | A |
| residue | 122 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 24) | chain | A |
| residue | 123 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 25) | chain | A |
| residue | 125 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 26) | chain | A |
| residue | 126 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 27) | chain | A |
| residue | 150 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 28) | chain | A |
| residue | 151 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 29) | chain | A |
| residue | 152 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GNP A 250 | |
| source | : AC3 | |
| 30) | chain | C |
| residue | 19 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 31) | chain | C |
| residue | 20 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 32) | chain | C |
| residue | 21 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 33) | chain | C |
| residue | 22 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 34) | chain | C |
| residue | 23 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 35) | chain | C |
| residue | 24 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 36) | chain | C |
| residue | 25 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 37) | chain | C |
| residue | 35 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 38) | chain | C |
| residue | 36 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 39) | chain | C |
| residue | 37 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 40) | chain | C |
| residue | 38 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 41) | chain | C |
| residue | 39 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 42) | chain | C |
| residue | 41 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 43) | chain | C |
| residue | 42 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 44) | chain | C |
| residue | 68 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 45) | chain | C |
| residue | 69 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 46) | chain | C |
| residue | 122 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 47) | chain | C |
| residue | 123 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 48) | chain | C |
| residue | 125 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 49) | chain | C |
| residue | 126 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 50) | chain | C |
| residue | 150 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 51) | chain | C |
| residue | 151 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 52) | chain | C |
| residue | 152 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GNP C 252 | |
| source | : AC4 | |
| 53) | chain | A |
| residue | 69 | |
| type | SITE | |
| sequence | Q | |
| description | Essential for GTP hydrolysis => ECO:0000269|PubMed:18591255, ECO:0000269|PubMed:26272610, ECO:0000269|PubMed:8636225 | |
| source | Swiss-Prot : SWS_FT_FI6 | |
| 54) | chain | C |
| residue | 69 | |
| type | SITE | |
| sequence | Q | |
| description | Essential for GTP hydrolysis => ECO:0000269|PubMed:18591255, ECO:0000269|PubMed:26272610, ECO:0000269|PubMed:8636225 | |
| source | Swiss-Prot : SWS_FT_FI6 | |
| 55) | chain | A |
| residue | 24 | |
| type | MOD_RES | |
| sequence | T | |
| description | Phosphothreonine => ECO:0007744|PubMed:23186163 | |
| source | Swiss-Prot : SWS_FT_FI8 | |
| 56) | chain | C |
| residue | 24 | |
| type | MOD_RES | |
| sequence | T | |
| description | Phosphothreonine => ECO:0007744|PubMed:23186163 | |
| source | Swiss-Prot : SWS_FT_FI8 | |
| 57) | chain | A |
| residue | 37 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine => ECO:0000269|PubMed:31075303 | |
| source | Swiss-Prot : SWS_FT_FI9 | |
| 58) | chain | C |
| residue | 37 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine => ECO:0000269|PubMed:31075303 | |
| source | Swiss-Prot : SWS_FT_FI9 | |
| 59) | chain | A |
| residue | 60 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine => ECO:0007744|PubMed:19608861 | |
| source | Swiss-Prot : SWS_FT_FI10 | |
| 60) | chain | A |
| residue | 99 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine => ECO:0007744|PubMed:19608861 | |
| source | Swiss-Prot : SWS_FT_FI10 | |
| 61) | chain | C |
| residue | 60 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine => ECO:0007744|PubMed:19608861 | |
| source | Swiss-Prot : SWS_FT_FI10 | |
| 62) | chain | C |
| residue | 99 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine => ECO:0007744|PubMed:19608861 | |
| source | Swiss-Prot : SWS_FT_FI10 | |
| 63) | chain | A |
| residue | 71 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine; alternate => ECO:0007744|PubMed:19608861 | |
| source | Swiss-Prot : SWS_FT_FI11 | |
| 64) | chain | C |
| residue | 71 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine; alternate => ECO:0007744|PubMed:19608861 | |
| source | Swiss-Prot : SWS_FT_FI11 | |
| 65) | chain | A |
| residue | 134 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine => ECO:0000269|PubMed:29040603 | |
| source | Swiss-Prot : SWS_FT_FI12 | |
| 66) | chain | C |
| residue | 134 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine => ECO:0000269|PubMed:29040603 | |
| source | Swiss-Prot : SWS_FT_FI12 | |
| 67) | chain | A |
| residue | 159 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine; alternate => ECO:0000250|UniProtKB:P62827 | |
| source | Swiss-Prot : SWS_FT_FI13 | |
| 68) | chain | C |
| residue | 159 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine; alternate => ECO:0000250|UniProtKB:P62827 | |
| source | Swiss-Prot : SWS_FT_FI13 | |
| 69) | chain | A |
| residue | 71 | |
| type | CROSSLNK | |
| sequence | K | |
| description | Glycyl lysine isopeptide (Lys-Gly) (interchain with G-Cter in ubiquitin); alternate | |
| source | Swiss-Prot : SWS_FT_FI14 | |
| 70) | chain | C |
| residue | 71 | |
| type | CROSSLNK | |
| sequence | K | |
| description | Glycyl lysine isopeptide (Lys-Gly) (interchain with G-Cter in ubiquitin); alternate | |
| source | Swiss-Prot : SWS_FT_FI14 | |
| 71) | chain | A |
| residue | 152 | |
| type | CROSSLNK | |
| sequence | K | |
| description | Glycyl lysine isopeptide (Lys-Gly) (interchain with G-Cter in SUMO2) => ECO:0007744|PubMed:28112733 | |
| source | Swiss-Prot : SWS_FT_FI15 | |
| 72) | chain | C |
| residue | 152 | |
| type | CROSSLNK | |
| sequence | K | |
| description | Glycyl lysine isopeptide (Lys-Gly) (interchain with G-Cter in SUMO2) => ECO:0007744|PubMed:28112733 | |
| source | Swiss-Prot : SWS_FT_FI15 | |
| 73) | chain | B |
| residue | 12 | |
| type | MOD_RES | |
| sequence | S | |
| description | Phosphoserine => ECO:0007744|PubMed:18669648, ECO:0007744|PubMed:23186163 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 74) | chain | D |
| residue | 12 | |
| type | MOD_RES | |
| sequence | S | |
| description | Phosphoserine => ECO:0007744|PubMed:18669648, ECO:0007744|PubMed:23186163 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 75) | chain | B |
| residue | 211 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine => ECO:0007744|PubMed:19608861 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 76) | chain | D |
| residue | 211 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine => ECO:0007744|PubMed:19608861 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 77) | chain | A |
| residue | 122 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000269|PubMed:10078529, ECO:0000269|PubMed:10353245, ECO:0000269|PubMed:10367892, ECO:0000269|PubMed:11832950, ECO:0000269|PubMed:18611384, ECO:0000269|PubMed:19389996, ECO:0000269|PubMed:19505478, ECO:0000269|PubMed:24449914, ECO:0000269|PubMed:24915079, ECO:0000269|PubMed:26272610, ECO:0000269|PubMed:26349033, ECO:0000269|PubMed:28282025, ECO:0000269|PubMed:7885480, ECO:0007744|PDB:1IBR, ECO:0007744|PDB:1K5D, ECO:0007744|PDB:1QBK, ECO:0007744|PDB:1RRP, ECO:0007744|PDB:3CH5, ECO:0007744|PDB:3GJ0, ECO:0007744|PDB:3GJ3, ECO:0007744|PDB:3GJ4, ECO:0007744|PDB:3GJ5, ECO:0007744|PDB:3GJ6, ECO:0007744|PDB:3GJ7, ECO:0007744|PDB:3GJ8, ECO:0007744|PDB:3GJX, ECO:0007744|PDB:3NBY, ECO:0007744|PDB:3NBZ, ECO:0007744|PDB:3NC0, ECO:0007744|PDB:3NC1, ECO:0007744|PDB:3ZJY, ECO:0007744|PDB:4C0Q, ECO:0007744|PDB:4OL0, ECO:0007744|PDB:5CIQ, ECO:0007744|PDB:5CIT, ECO:0007744|PDB:5CIW, ECO:0007744|PDB:5CJ2, ECO:0007744|PDB:5CLL, ECO:0007744|PDB:5DH9, ECO:0007744|PDB:5DHA, ECO:0007744|PDB:5DHF, ECO:0007744|PDB:5DI9, ECO:0007744|PDB:5DIF, ECO:0007744|PDB:5DIS, ECO:0007744|PDB:5DLQ, ECO:0007744|PDB:5JLJ, ECO:0007744|PDB:5UWH, ECO:0007744|PDB:5UWI, ECO:0007744|PDB:5UWJ, ECO:0007744|PDB:5UWO, ECO:0007744|PDB:5UWP, ECO:0007744|PDB:5UWQ, ECO:0007744|PDB:5UWR, ECO:0007744|PDB:5UWS, ECO:0007744|PDB:5UWT, ECO:0007744|PDB:5UWU, ECO:0007744|PDB:5UWW | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 78) | chain | C |
| residue | 122 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000269|PubMed:10078529, ECO:0000269|PubMed:10353245, ECO:0000269|PubMed:10367892, ECO:0000269|PubMed:11832950, ECO:0000269|PubMed:18611384, ECO:0000269|PubMed:19389996, ECO:0000269|PubMed:19505478, ECO:0000269|PubMed:24449914, ECO:0000269|PubMed:24915079, ECO:0000269|PubMed:26272610, ECO:0000269|PubMed:26349033, ECO:0000269|PubMed:28282025, ECO:0000269|PubMed:7885480, ECO:0007744|PDB:1IBR, ECO:0007744|PDB:1K5D, ECO:0007744|PDB:1QBK, ECO:0007744|PDB:1RRP, ECO:0007744|PDB:3CH5, ECO:0007744|PDB:3GJ0, ECO:0007744|PDB:3GJ3, ECO:0007744|PDB:3GJ4, ECO:0007744|PDB:3GJ5, ECO:0007744|PDB:3GJ6, ECO:0007744|PDB:3GJ7, ECO:0007744|PDB:3GJ8, ECO:0007744|PDB:3GJX, ECO:0007744|PDB:3NBY, ECO:0007744|PDB:3NBZ, ECO:0007744|PDB:3NC0, ECO:0007744|PDB:3NC1, ECO:0007744|PDB:3ZJY, ECO:0007744|PDB:4C0Q, ECO:0007744|PDB:4OL0, ECO:0007744|PDB:5CIQ, ECO:0007744|PDB:5CIT, ECO:0007744|PDB:5CIW, ECO:0007744|PDB:5CJ2, ECO:0007744|PDB:5CLL, ECO:0007744|PDB:5DH9, ECO:0007744|PDB:5DHA, ECO:0007744|PDB:5DHF, ECO:0007744|PDB:5DI9, ECO:0007744|PDB:5DIF, ECO:0007744|PDB:5DIS, ECO:0007744|PDB:5DLQ, ECO:0007744|PDB:5JLJ, ECO:0007744|PDB:5UWH, ECO:0007744|PDB:5UWI, ECO:0007744|PDB:5UWJ, ECO:0007744|PDB:5UWO, ECO:0007744|PDB:5UWP, ECO:0007744|PDB:5UWQ, ECO:0007744|PDB:5UWR, ECO:0007744|PDB:5UWS, ECO:0007744|PDB:5UWT, ECO:0007744|PDB:5UWU, ECO:0007744|PDB:5UWW | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 79) | chain | A |
| residue | 150 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000269|PubMed:10078529, ECO:0000269|PubMed:10353245, ECO:0000269|PubMed:10367892, ECO:0000269|PubMed:11832950, ECO:0000269|PubMed:18611384, ECO:0000269|PubMed:19389996, ECO:0000269|PubMed:19505478, ECO:0000269|PubMed:24449914, ECO:0000269|PubMed:24915079, ECO:0000269|PubMed:26272610, ECO:0000269|PubMed:26349033, ECO:0000269|PubMed:28282025, ECO:0000269|PubMed:7885480, ECO:0007744|PDB:1K5G, ECO:0007744|PDB:3CH5, ECO:0007744|PDB:3GJ0, ECO:0007744|PDB:3GJ3, ECO:0007744|PDB:3GJ4, ECO:0007744|PDB:3GJ5, ECO:0007744|PDB:3GJ6, ECO:0007744|PDB:3GJ7, ECO:0007744|PDB:3GJ8, ECO:0007744|PDB:3GJX, ECO:0007744|PDB:3NBY, ECO:0007744|PDB:3NBZ, ECO:0007744|PDB:3NC0, ECO:0007744|PDB:3NC1, ECO:0007744|PDB:3ZJY, ECO:0007744|PDB:4C0Q, ECO:0007744|PDB:4OL0, ECO:0007744|PDB:5CIQ, ECO:0007744|PDB:5CIT, ECO:0007744|PDB:5CIW, ECO:0007744|PDB:5CJ2, ECO:0007744|PDB:5CLL, ECO:0007744|PDB:5CLQ, ECO:0007744|PDB:5DH9, ECO:0007744|PDB:5DHA, ECO:0007744|PDB:5DHF, ECO:0007744|PDB:5DI9, ECO:0007744|PDB:5DIF, ECO:0007744|PDB:5DIS, ECO:0007744|PDB:5DLQ, ECO:0007744|PDB:5JLJ, ECO:0007744|PDB:5UWH, ECO:0007744|PDB:5UWI, ECO:0007744|PDB:5UWJ, ECO:0007744|PDB:5UWO, ECO:0007744|PDB:5UWP, ECO:0007744|PDB:5UWQ, ECO:0007744|PDB:5UWR, ECO:0007744|PDB:5UWS, ECO:0007744|PDB:5UWT, ECO:0007744|PDB:5UWU, ECO:0007744|PDB:5UWW | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 80) | chain | C |
| residue | 150 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000269|PubMed:10078529, ECO:0000269|PubMed:10353245, ECO:0000269|PubMed:10367892, ECO:0000269|PubMed:11832950, ECO:0000269|PubMed:18611384, ECO:0000269|PubMed:19389996, ECO:0000269|PubMed:19505478, ECO:0000269|PubMed:24449914, ECO:0000269|PubMed:24915079, ECO:0000269|PubMed:26272610, ECO:0000269|PubMed:26349033, ECO:0000269|PubMed:28282025, ECO:0000269|PubMed:7885480, ECO:0007744|PDB:1K5G, ECO:0007744|PDB:3CH5, ECO:0007744|PDB:3GJ0, ECO:0007744|PDB:3GJ3, ECO:0007744|PDB:3GJ4, ECO:0007744|PDB:3GJ5, ECO:0007744|PDB:3GJ6, ECO:0007744|PDB:3GJ7, ECO:0007744|PDB:3GJ8, ECO:0007744|PDB:3GJX, ECO:0007744|PDB:3NBY, ECO:0007744|PDB:3NBZ, ECO:0007744|PDB:3NC0, ECO:0007744|PDB:3NC1, ECO:0007744|PDB:3ZJY, ECO:0007744|PDB:4C0Q, ECO:0007744|PDB:4OL0, ECO:0007744|PDB:5CIQ, ECO:0007744|PDB:5CIT, ECO:0007744|PDB:5CIW, ECO:0007744|PDB:5CJ2, ECO:0007744|PDB:5CLL, ECO:0007744|PDB:5CLQ, ECO:0007744|PDB:5DH9, ECO:0007744|PDB:5DHA, ECO:0007744|PDB:5DHF, ECO:0007744|PDB:5DI9, ECO:0007744|PDB:5DIF, ECO:0007744|PDB:5DIS, ECO:0007744|PDB:5DLQ, ECO:0007744|PDB:5JLJ, ECO:0007744|PDB:5UWH, ECO:0007744|PDB:5UWI, ECO:0007744|PDB:5UWJ, ECO:0007744|PDB:5UWO, ECO:0007744|PDB:5UWP, ECO:0007744|PDB:5UWQ, ECO:0007744|PDB:5UWR, ECO:0007744|PDB:5UWS, ECO:0007744|PDB:5UWT, ECO:0007744|PDB:5UWU, ECO:0007744|PDB:5UWW | |
| source | Swiss-Prot : SWS_FT_FI5 | |