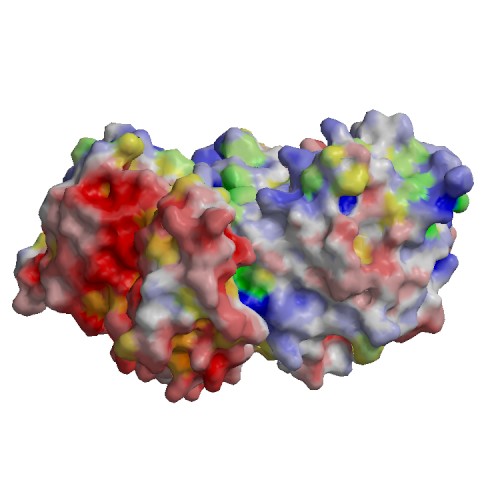
Functional site
| 1) | chain | B |
| residue | 1039 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 2) | chain | B |
| residue | 1074 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 3) | chain | B |
| residue | 1076 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 4) | chain | B |
| residue | 1077 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 5) | chain | B |
| residue | 1078 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 6) | chain | B |
| residue | 1079 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 7) | chain | B |
| residue | 1080 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 8) | chain | B |
| residue | 1081 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 9) | chain | B |
| residue | 1084 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 10) | chain | B |
| residue | 1085 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 11) | chain | B |
| residue | 1142 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 12) | chain | B |
| residue | 1143 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 13) | chain | B |
| residue | 1144 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 14) | chain | B |
| residue | 1150 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 15) | chain | B |
| residue | 1152 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 16) | chain | B |
| residue | 1153 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 17) | chain | B |
| residue | 1156 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 18) | chain | B |
| residue | 1159 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 19) | chain | B |
| residue | 1160 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 20) | chain | B |
| residue | 1257 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 21) | chain | B |
| residue | 1258 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 22) | chain | B |
| residue | 1260 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 23) | chain | B |
| residue | 1261 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 24) | chain | B |
| residue | 1262 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 25) | chain | B |
| residue | 1528 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 26) | chain | B |
| residue | 1529 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 27) | chain | B |
| residue | 1555 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE FAD B 1600 | |
| source | : AC2 | |
| 28) | chain | B |
| residue | 1150 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000255|HAMAP-Rule:MF_02092, ECO:0000269|PubMed:10944213, ECO:0007744|PDB:1F0X | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 29) | chain | B |
| residue | 1160 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000255|HAMAP-Rule:MF_02092, ECO:0000269|PubMed:10944213, ECO:0007744|PDB:1F0X | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 30) | chain | B |
| residue | 1262 | |
| type | BINDING | |
| sequence | V | |
| description | BINDING => ECO:0000255|HAMAP-Rule:MF_02092, ECO:0000269|PubMed:10944213, ECO:0007744|PDB:1F0X | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 31) | chain | B |
| residue | 1076 | |
| type | BINDING | |
| sequence | A | |
| description | BINDING => ECO:0000255|HAMAP-Rule:MF_02092, ECO:0000269|PubMed:10944213, ECO:0007744|PDB:1F0X | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 32) | chain | B |
| residue | 1084 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000255|HAMAP-Rule:MF_02092, ECO:0000269|PubMed:10944213, ECO:0007744|PDB:1F0X | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 33) | chain | B |
| residue | 1143 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000255|HAMAP-Rule:MF_02092, ECO:0000269|PubMed:10944213, ECO:0007744|PDB:1F0X | |
| source | Swiss-Prot : SWS_FT_FI1 | |