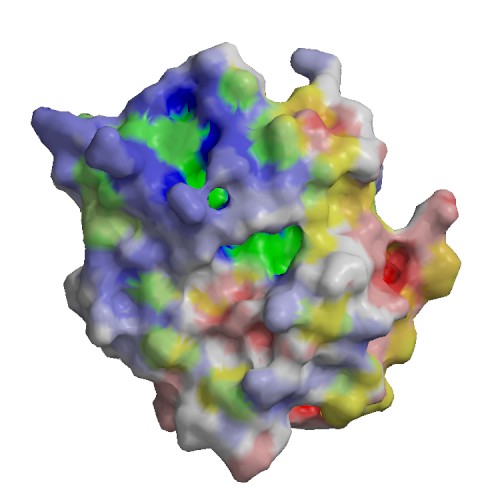
Functional site
| 1) | chain | A |
| residue | 145 | |
| type | ||
| sequence | X | |
| description | ACTIVE SITE CYSTEINE, BENZYLATED | |
| source | : ACT | |
| 2) | chain | A |
| residue | 5 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN A 208 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 24 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN A 208 | |
| source | : AC1 | |
| 4) | chain | A |
| residue | 29 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN A 208 | |
| source | : AC1 | |
| 5) | chain | A |
| residue | 85 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN A 208 | |
| source | : AC1 | |
| 6) | chain | A |
| residue | 114 | |
| type | catalytic | |
| sequence | Y | |
| description | 251 | |
| source | MCSA : MCSA1 | |
| 7) | chain | A |
| residue | 137 | |
| type | catalytic | |
| sequence | N | |
| description | 251 | |
| source | MCSA : MCSA1 | |
| 8) | chain | A |
| residue | 145 | |
| type | catalytic | |
| sequence | X | |
| description | 251 | |
| source | MCSA : MCSA1 | |
| 9) | chain | A |
| residue | 146 | |
| type | catalytic | |
| sequence | H | |
| description | 251 | |
| source | MCSA : MCSA1 | |
| 10) | chain | A |
| residue | 172 | |
| type | catalytic | |
| sequence | E | |
| description | 251 | |
| source | MCSA : MCSA1 | |
| 11) | chain | A |
| residue | 143-149 | |
| type | prosite | |
| sequence | IPXHRVV | |
| description | MGMT Methylated-DNA--protein-cysteine methyltransferase active site. IPCHRVV | |
| source | prosite : PS00374 | |
| 12) | chain | A |
| residue | 123 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000269|PubMed:10747039, ECO:0000269|PubMed:15964013 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 13) | chain | A |
| residue | 128 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000269|PubMed:10747039, ECO:0000269|PubMed:15964013 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 14) | chain | A |
| residue | 151 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000269|PubMed:10747039, ECO:0000269|PubMed:15964013 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 15) | chain | A |
| residue | 95 | |
| type | BINDING | |
| sequence | T | |
| description | BINDING => ECO:0000269|PubMed:10747039, ECO:0000269|PubMed:15964013 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 16) | chain | A |
| residue | 114 | |
| type | BINDING | |
| sequence | Y | |
| description | BINDING => ECO:0000269|PubMed:10747039, ECO:0000269|PubMed:15964013 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 17) | chain | A |
| residue | 115 | |
| type | BINDING | |
| sequence | Q | |
| description | BINDING => ECO:0000269|PubMed:10747039, ECO:0000269|PubMed:15964013 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 18) | chain | A |
| residue | 14 | |
| type | MOD_RES | |
| sequence | S | |
| description | Phosphoserine => ECO:0007744|PubMed:23186163 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 19) | chain | A |
| residue | 145 | |
| type | ACT_SITE | |
| sequence | X | |
| description | Nucleophile; methyl group acceptor | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 20) | chain | A |
| residue | 5 | |
| type | BINDING | |
| sequence | C | |
| description | ||
| source | Swiss-Prot : SWS_FT_FI2 | |
| 21) | chain | A |
| residue | 24 | |
| type | BINDING | |
| sequence | C | |
| description | ||
| source | Swiss-Prot : SWS_FT_FI2 | |
| 22) | chain | A |
| residue | 29 | |
| type | BINDING | |
| sequence | H | |
| description | ||
| source | Swiss-Prot : SWS_FT_FI2 | |
| 23) | chain | A |
| residue | 85 | |
| type | BINDING | |
| sequence | H | |
| description | ||
| source | Swiss-Prot : SWS_FT_FI2 | |