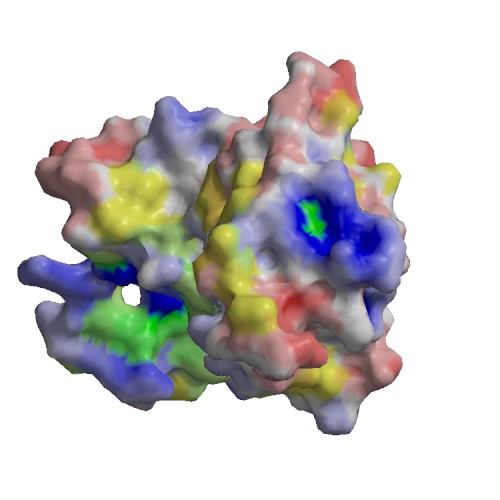
Functional site
| 1) | chain | A |
| residue | 23 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE PO4 A 275 | |
| source | : AC1 | |
| 2) | chain | A |
| residue | 24 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE PO4 A 275 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 120 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE PO4 A 275 | |
| source | : AC1 | |
| 4) | chain | A |
| residue | 152 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE PO4 A 275 | |
| source | : AC1 | |
| 5) | chain | A |
| residue | 153 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE PO4 A 275 | |
| source | : AC1 | |
| 6) | chain | A |
| residue | 215 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE PO4 A 275 | |
| source | : AC1 | |
| 7) | chain | A |
| residue | 67 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 8) | chain | A |
| residue | 68 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 9) | chain | A |
| residue | 69 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 10) | chain | A |
| residue | 70 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 11) | chain | A |
| residue | 82 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 12) | chain | A |
| residue | 83 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 13) | chain | A |
| residue | 87 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 14) | chain | A |
| residue | 88 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 15) | chain | A |
| residue | 89 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 16) | chain | A |
| residue | 111 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 17) | chain | A |
| residue | 146 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 18) | chain | A |
| residue | 147 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 19) | chain | A |
| residue | 191 | |
| type | ||
| sequence | W | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 20) | chain | A |
| residue | 201 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE FMN A 250 | |
| source | : AC2 | |
| 21) | chain | A |
| residue | 198 | |
| type | catalytic | |
| sequence | L | |
| description | 677 | |
| source | MCSA : MCSA1 | |
| 22) | chain | A |
| residue | 192 | |
| type | BINDING | |
| sequence | Q | |
| description | BINDING => ECO:0000269|PubMed:15858270 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 23) | chain | A |
| residue | 198 | |
| type | BINDING | |
| sequence | L | |
| description | BINDING => ECO:0000269|PubMed:11786019 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 24) | chain | A |
| residue | 68 | |
| type | BINDING | |
| sequence | I | |
| description | BINDING => ECO:0000269|PubMed:10903950, ECO:0000269|PubMed:11453690, ECO:0000269|PubMed:15858270 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 25) | chain | A |
| residue | 83 | |
| type | BINDING | |
| sequence | T | |
| description | BINDING => ECO:0000269|PubMed:10903950, ECO:0000269|PubMed:11453690, ECO:0000269|PubMed:15858270 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 26) | chain | A |
| residue | 89 | |
| type | BINDING | |
| sequence | K | |
| description | BINDING => ECO:0000269|PubMed:10903950, ECO:0000269|PubMed:11453690, ECO:0000269|PubMed:15858270 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 27) | chain | A |
| residue | 90 | |
| type | BINDING | |
| sequence | A | |
| description | BINDING => ECO:0000269|PubMed:10903950, ECO:0000269|PubMed:11453690, ECO:0000269|PubMed:15858270 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 28) | chain | A |
| residue | 147 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000269|PubMed:10903950, ECO:0000269|PubMed:11453690, ECO:0000269|PubMed:15858270 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 29) | chain | A |
| residue | 73 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000269|PubMed:11453690 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 30) | chain | A |
| residue | 130 | |
| type | BINDING | |
| sequence | F | |
| description | BINDING => ECO:0000269|PubMed:11453690 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 31) | chain | A |
| residue | 134 | |
| type | BINDING | |
| sequence | P | |
| description | BINDING => ECO:0000269|PubMed:11453690 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 32) | chain | A |
| residue | 138 | |
| type | BINDING | |
| sequence | Q | |
| description | BINDING => ECO:0000269|PubMed:11453690 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 33) | chain | A |
| residue | 112 | |
| type | BINDING | |
| sequence | V | |
| description | BINDING => ECO:0000269|PubMed:11786019, ECO:0000269|PubMed:15858270 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 34) | chain | A |
| residue | 202 | |
| type | BINDING | |
| sequence | F | |
| description | BINDING => ECO:0000269|PubMed:11786019, ECO:0000269|PubMed:15858270 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 35) | chain | A |
| residue | 188-201 | |
| type | prosite | |
| sequence | IEFWQGGEHRLHDR | |
| description | PYRIDOX_OXIDASE Pyridoxamine 5'-phosphate oxidase signature. IEFWQggehRLHDR | |
| source | prosite : PS01064 | |