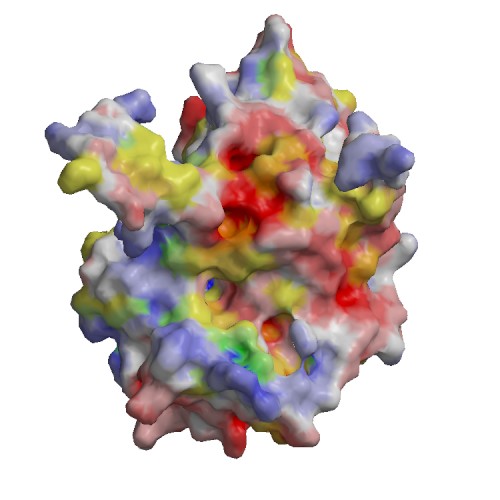
Functional site
| 1) | chain | A |
| residue | 260 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE SO4 A 808 | |
| source | : AC2 | |
| 2) | chain | A |
| residue | 263 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE SO4 A 808 | |
| source | : AC2 | |
| 3) | chain | A |
| residue | 266 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 A 808 | |
| source | : AC2 | |
| 4) | chain | A |
| residue | 298 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE SO4 A 810 | |
| source | : AC4 | |
| 5) | chain | A |
| residue | 302 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 A 810 | |
| source | : AC4 | |
| 6) | chain | A |
| residue | 41 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 A 811 | |
| source | : AC5 | |
| 7) | chain | A |
| residue | 199 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE SO4 A 811 | |
| source | : AC5 | |
| 8) | chain | A |
| residue | 200 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE SO4 A 811 | |
| source | : AC5 | |
| 9) | chain | A |
| residue | 89 | |
| type | ||
| sequence | X | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 10) | chain | A |
| residue | 148 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 11) | chain | A |
| residue | 156 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 12) | chain | A |
| residue | 157 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 13) | chain | A |
| residue | 220 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 14) | chain | A |
| residue | 227 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 15) | chain | A |
| residue | 228 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 16) | chain | A |
| residue | 243 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 17) | chain | A |
| residue | 244 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 18) | chain | A |
| residue | 247 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 19) | chain | A |
| residue | 248 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 20) | chain | A |
| residue | 288 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 21) | chain | A |
| residue | 318 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 22) | chain | A |
| residue | 319 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 23) | chain | A |
| residue | 348 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 24) | chain | A |
| residue | 378 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ACO A 813 | |
| source | : AC7 | |
| 25) | chain | A |
| residue | 85-103 | |
| type | prosite | |
| sequence | MNQLXGSGLRAVALGMQQI | |
| description | THIOLASE_1 Thiolases acyl-enzyme intermediate signature. MNQlCGSGLrAValgmqqI | |
| source | prosite : PS00098 | |
| 26) | chain | A |
| residue | 373-386 | |
| type | prosite | |
| sequence | GLATLCIGGGMGVA | |
| description | THIOLASE_3 Thiolases active site. GLATLCIGgGmGvA | |
| source | prosite : PS00099 | |
| 27) | chain | A |
| residue | 338-354 | |
| type | prosite | |
| sequence | NVNGGAIAIGHPIGASG | |
| description | THIOLASE_2 Thiolases signature 2. NvnGGaIAiGHPiGaSG | |
| source | prosite : PS00737 | |
| 28) | chain | A |
| residue | 89 | |
| type | ACT_SITE | |
| sequence | X | |
| description | Acyl-thioester intermediate | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 29) | chain | A |
| residue | 348 | |
| type | ACT_SITE | |
| sequence | H | |
| description | Proton acceptor | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 30) | chain | A |
| residue | 378 | |
| type | ACT_SITE | |
| sequence | C | |
| description | Proton acceptor | |
| source | Swiss-Prot : SWS_FT_FI2 | |