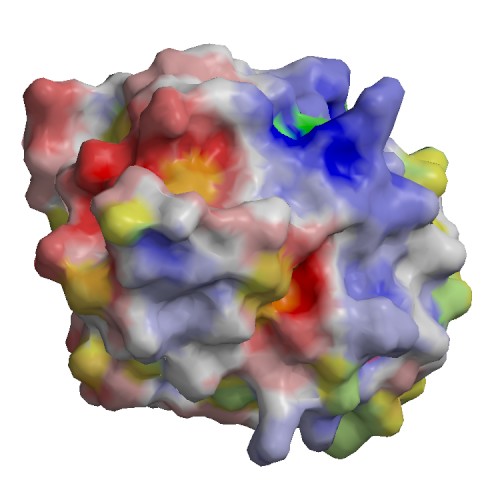
Functional site
| 1) | chain | A |
| residue | 120 | |
| type | ||
| sequence | S | |
| description | CATALYTIC TRIAD | |
| source | : CAT | |
| 2) | chain | A |
| residue | 188 | |
| type | ||
| sequence | H | |
| description | CATALYTIC TRIAD | |
| source | : CAT | |
| 3) | chain | A |
| residue | 175 | |
| type | ||
| sequence | D | |
| description | CATALYTIC TRIAD | |
| source | : CAT | |
| 4) | chain | A |
| residue | 120 | |
| type | ACT_SITE | |
| sequence | S | |
| description | Nucleophile => ECO:0000269|PubMed:1560844, ECO:0007744|PDB:1AGY | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 5) | chain | A |
| residue | 42 | |
| type | catalytic | |
| sequence | S | |
| description | 631 | |
| source | MCSA : MCSA1 | |
| 6) | chain | A |
| residue | 120 | |
| type | catalytic | |
| sequence | S | |
| description | 631 | |
| source | MCSA : MCSA1 | |
| 7) | chain | A |
| residue | 121 | |
| type | catalytic | |
| sequence | Q | |
| description | 631 | |
| source | MCSA : MCSA1 | |
| 8) | chain | A |
| residue | 175 | |
| type | catalytic | |
| sequence | D | |
| description | 631 | |
| source | MCSA : MCSA1 | |
| 9) | chain | A |
| residue | 188 | |
| type | catalytic | |
| sequence | H | |
| description | 631 | |
| source | MCSA : MCSA1 | |
| 10) | chain | A |
| residue | 110-122 | |
| type | prosite | |
| sequence | PDATLIAGGYSQG | |
| description | CUTINASE_1 Cutinase, serine active site. PdAtLIaGGYSQG | |
| source | prosite : PS00155 | |
| 11) | chain | A |
| residue | 171-188 | |
| type | prosite | |
| sequence | CKTGDLVCTGSLIVAAPH | |
| description | CUTINASE_2 Cutinase, aspartate and histidine active sites. CktgDlVCtGSliVaapH | |
| source | prosite : PS00931 | |
| 12) | chain | A |
| residue | 188 | |
| type | ACT_SITE | |
| sequence | H | |
| description | Proton donor/acceptor => ECO:0000269|PubMed:1560844, ECO:0007744|PDB:1AGY | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 13) | chain | A |
| residue | 42 | |
| type | SITE | |
| sequence | S | |
| description | Transition state stabilizer => ECO:0000269|PubMed:1560844, ECO:0000269|PubMed:8286366, ECO:0000269|PubMed:8555209, ECO:0000269|PubMed:9041628, ECO:0007744|PDB:1AGY, ECO:0007744|PDB:1FFE, ECO:0007744|PDB:1OXM, ECO:0007744|PDB:2CUT | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 14) | chain | A |
| residue | 121 | |
| type | SITE | |
| sequence | Q | |
| description | Transition state stabilizer => ECO:0000269|PubMed:1560844, ECO:0000269|PubMed:8286366, ECO:0000269|PubMed:9041628, ECO:0007744|PDB:1AGY, ECO:0007744|PDB:1OXM, ECO:0007744|PDB:2CUT | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 15) | chain | A |
| residue | 175 | |
| type | ACT_SITE | |
| sequence | D | |
| description | ACT_SITE => ECO:0000269|PubMed:1560844, ECO:0007744|PDB:1AGY | |
| source | Swiss-Prot : SWS_FT_FI2 | |