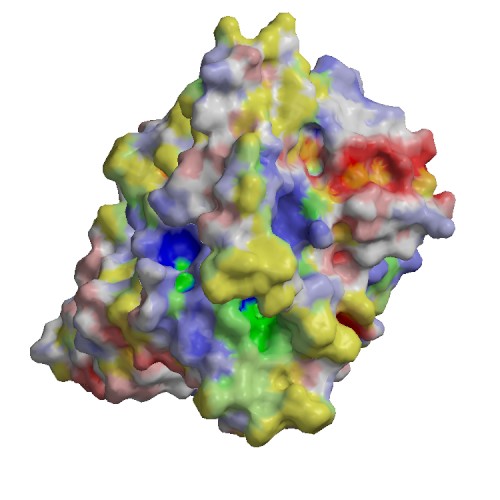
Functional site
| 1) | chain | B |
| residue | 120 | |
| type | ||
| sequence | R | |
| description | HYDROPHOBIC CHANNEL RESPONSIBLE FOR CYCLOOGENASE ACTIVITY | |
| source | : COB | |
| 2) | chain | B |
| residue | 355 | |
| type | ||
| sequence | Y | |
| description | HYDROPHOBIC CHANNEL RESPONSIBLE FOR CYCLOOGENASE ACTIVITY | |
| source | : COB | |
| 3) | chain | B |
| residue | 385 | |
| type | ||
| sequence | Y | |
| description | HYDROPHOBIC CHANNEL RESPONSIBLE FOR CYCLOOGENASE ACTIVITY | |
| source | : COB | |
| 4) | chain | B |
| residue | 387 | |
| type | ||
| sequence | W | |
| description | HYDROPHOBIC CHANNEL RESPONSIBLE FOR CYCLOOGENASE ACTIVITY | |
| source | : COB | |
| 5) | chain | B |
| residue | 523 | |
| type | ||
| sequence | I | |
| description | HYDROPHOBIC CHANNEL RESPONSIBLE FOR CYCLOOGENASE ACTIVITY | |
| source | : COB | |
| 6) | chain | B |
| residue | 530 | |
| type | ||
| sequence | S | |
| description | HYDROPHOBIC CHANNEL RESPONSIBLE FOR CYCLOOGENASE ACTIVITY | |
| source | : COB | |
| 7) | chain | B |
| residue | 207 | |
| type | ||
| sequence | H | |
| description | HEME POCKET RESPONSIBLE FOR PEROXIDASE ACTIVITY | |
| source | : POB | |
| 8) | chain | B |
| residue | 388 | |
| type | ||
| sequence | H | |
| description | HEME POCKET RESPONSIBLE FOR PEROXIDASE ACTIVITY | |
| source | : POB | |
| 9) | chain | B |
| residue | 74-82 | |
| type | TRANSMEM | |
| sequence | IWTWLRTTL | |
| description | Helical | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 10) | chain | B |
| residue | 86-92 | |
| type | TRANSMEM | |
| sequence | PSFIHFL | |
| description | Helical | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 11) | chain | B |
| residue | 97-105 | |
| type | TRANSMEM | |
| sequence | RWLWDFVNA | |
| description | Helical | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 12) | chain | B |
| residue | 108-122 | |
| type | TRANSMEM | |
| sequence | IRDTLMRLVLTVRSN | |
| description | Helical | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 13) | chain | B |
| residue | 207 | |
| type | ACT_SITE | |
| sequence | H | |
| description | Proton acceptor | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 14) | chain | B |
| residue | 385 | |
| type | ACT_SITE | |
| sequence | Y | |
| description | For cyclooxygenase activity => ECO:0000269|PubMed:2122967 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 15) | chain | B |
| residue | 388 | |
| type | BINDING | |
| sequence | H | |
| description | axial binding residue | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 16) | chain | B |
| residue | 104 | |
| type | SITE | |
| sequence | N | |
| description | Not glycosylated => ECO:0000269|PubMed:8349699 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 17) | chain | B |
| residue | 530 | |
| type | SITE | |
| sequence | S | |
| description | Aspirin-acetylated serine | |
| source | Swiss-Prot : SWS_FT_FI6 | |
| 18) | chain | B |
| residue | 68 | |
| type | CARBOHYD | |
| sequence | N | |
| description | N-linked (GlcNAc...) asparagine => ECO:0000269|PubMed:8349699 | |
| source | Swiss-Prot : SWS_FT_FI7 | |
| 19) | chain | B |
| residue | 144 | |
| type | CARBOHYD | |
| sequence | N | |
| description | N-linked (GlcNAc...) asparagine => ECO:0000269|PubMed:8349699 | |
| source | Swiss-Prot : SWS_FT_FI7 | |
| 20) | chain | B |
| residue | 410 | |
| type | CARBOHYD | |
| sequence | N | |
| description | N-linked (GlcNAc...) asparagine => ECO:0000269|PubMed:8349699 | |
| source | Swiss-Prot : SWS_FT_FI7 | |