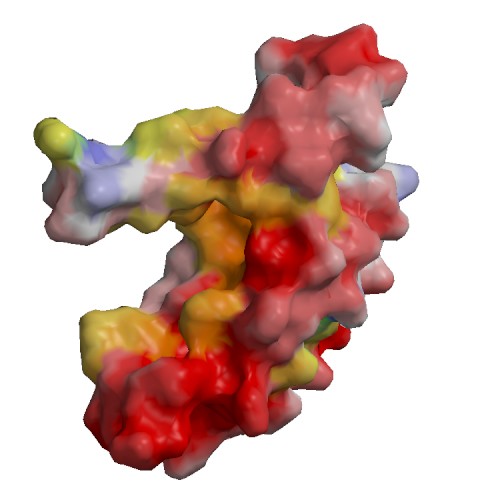
Functional site
| 1) | chain | A |
| residue | 2 | |
| type | MOD_RES | |
| sequence | E | |
| description | N-acetylserine => ECO:0000250|UniProtKB:P02638 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 2) | chain | A |
| residue | 62 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00448, ECO:0000269|PubMed:18949447, ECO:0007744|PDB:2K7O | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 3) | chain | A |
| residue | 66 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00448, ECO:0000269|PubMed:18949447, ECO:0007744|PDB:2K7O | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 4) | chain | A |
| residue | 68 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00448, ECO:0000269|PubMed:18949447, ECO:0007744|PDB:2K7O | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 5) | chain | A |
| residue | 73 | |
| type | BINDING | |
| sequence | F | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00448, ECO:0000269|PubMed:18949447, ECO:0007744|PDB:2K7O | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 6) | chain | A |
| residue | 64 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00448, ECO:0000269|PubMed:18949447, ECO:0007744|PDB:2K7O | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 7) | chain | A |
| residue | 61-73 | |
| type | prosite | |
| sequence | DEDGDGECDFQEF | |
| description | EF_HAND_1 EF-hand calcium-binding domain. DEDGDGECDfqEF | |
| source | prosite : PS00018 | |
| 8) | chain | A |
| residue | 56-77 | |
| type | prosite | |
| sequence | VMETLDEDGDGECDFQEFMAFV | |
| description | S100_CABP S-100/ICaBP type calcium binding protein signature. VMetLDedgDgecDFqEFmaFV | |
| source | prosite : PS00303 | |
| 9) | chain | A |
| residue | 16 | |
| type | BINDING | |
| sequence | Q | |
| description | BINDING => ECO:0000305|PubMed:14621986 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 10) | chain | A |
| residue | 26 | |
| type | BINDING | |
| sequence | K | |
| description | BINDING => ECO:0000305|PubMed:14621986 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 11) | chain | A |
| residue | 86 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000305|PubMed:14621986 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 12) | chain | A |
| residue | 91 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000305|PubMed:14621986 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 13) | chain | A |
| residue | 22 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000269|PubMed:18949447, ECO:0007744|PDB:2K7O | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 14) | chain | A |
| residue | 19 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000269|PubMed:18949447, ECO:0007744|PDB:2K7O | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 15) | chain | A |
| residue | 27 | |
| type | BINDING | |
| sequence | L | |
| description | BINDING => ECO:0000269|PubMed:18949447, ECO:0007744|PDB:2K7O | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 16) | chain | A |
| residue | 32 | |
| type | BINDING | |
| sequence | L | |
| description | BINDING => ECO:0000269|PubMed:18949447, ECO:0007744|PDB:2K7O | |
| source | Swiss-Prot : SWS_FT_FI2 | |