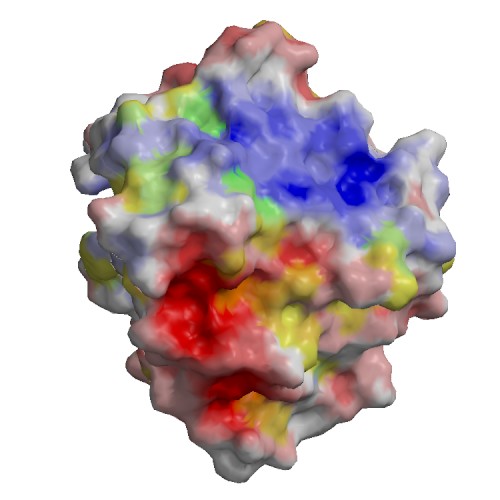
Functional site
| 1) | chain | D |
| residue | 39 | |
| type | catalytic | |
| sequence | E | |
| description | 41 | |
| source | MCSA : MCSA1 | |
| 2) | chain | D |
| residue | 76 | |
| type | catalytic | |
| sequence | Y | |
| description | 41 | |
| source | MCSA : MCSA1 | |
| 3) | chain | D |
| residue | 78 | |
| type | catalytic | |
| sequence | E | |
| description | 41 | |
| source | MCSA : MCSA1 | |
| 4) | chain | D |
| residue | 134 | |
| type | catalytic | |
| sequence | H | |
| description | 41 | |
| source | MCSA : MCSA1 | |
| 5) | chain | D |
| residue | 168 | |
| type | catalytic | |
| sequence | D | |
| description | 41 | |
| source | MCSA : MCSA1 | |
| 6) | chain | D |
| residue | 212 | |
| type | catalytic | |
| sequence | D | |
| description | 41 | |
| source | MCSA : MCSA1 | |
| 7) | chain | D |
| residue | 252 | |
| type | catalytic | |
| sequence | H | |
| description | 41 | |
| source | MCSA : MCSA1 | |
| 8) | chain | D |
| residue | 78 | |
| type | ACT_SITE | |
| sequence | E | |
| description | ACT_SITE => ECO:0000269|PubMed:2395459 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 9) | chain | D |
| residue | 134 | |
| type | ACT_SITE | |
| sequence | H | |
| description | ACT_SITE => ECO:0000269|PubMed:2395459, ECO:0000269|PubMed:4976790 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 10) | chain | D |
| residue | 67 | |
| type | SITE | |
| sequence | V | |
| description | Involved in actin-binding => ECO:0000269|PubMed:2395459 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 11) | chain | D |
| residue | 13 | |
| type | SITE | |
| sequence | E | |
| description | Involved in actin-binding => ECO:0000269|PubMed:2395459 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 12) | chain | D |
| residue | 65 | |
| type | SITE | |
| sequence | Y | |
| description | Nitration by tetranitromethane destroys a Ca(2+) binding site and inactivates enzyme | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 13) | chain | D |
| residue | 18 | |
| type | CARBOHYD | |
| sequence | N | |
| description | N-linked (GlcNAc...) asparagine => ECO:0000269|PubMed:1748997, ECO:0000269|PubMed:3560229 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 14) | chain | D |
| residue | 167-174 | |
| type | prosite | |
| sequence | GDFNADCS | |
| description | DNASE_I_2 Deoxyribonuclease I signature 2. GDFNAdCS | |
| source | prosite : PS00918 | |
| 15) | chain | D |
| residue | 130-150 | |
| type | prosite | |
| sequence | IVALHSAPSDAVAEINSLYDV | |
| description | DNASE_I_1 Deoxyribonuclease I signature 1. IVALHSAPsdavaEINsLyDV | |
| source | prosite : PS00919 | |