2WQ9
 
 | | Crystal Structure of RBP4 bound to Oleic Acid | | Descriptor: | CHLORIDE ION, GLYCEROL, OLEIC ACID, ... | | Authors: | Nanao, M, Mercer, D, Nguyen, L, Buckley, D, Stout, T.J. | | Deposit date: | 2009-08-14 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Rbp4 Bound to Oleic Acid
To be Published
|
|
2WQA
 
 | | Complex of TTR and RBP4 and Oleic Acid | | Descriptor: | OLEIC ACID, RETINOL-BINDING PROTEIN 4, SULFATE ION, ... | | Authors: | Nanao, M, Mercer, D, Nguyen, L, Buckley, D, Stout, T.J. | | Deposit date: | 2009-08-14 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Rbp4 Bound to Linoleic Acid and Ttr
To be Published
|
|
4RAK
 
 | |
2XHY
 
 | | Crystal Structure of E.coli BglA | | Descriptor: | 6-PHOSPHO-BETA-GLUCOSIDASE BGLA, BROMIDE ION, SULFATE ION | | Authors: | Totir, M, Zubieta, C, Echols, N, May, A.P, Gee, C.L, nanao, M, alber, T. | | Deposit date: | 2010-06-24 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
6H2M
 
 | | Long wavelength Mesh&Collect native SAD phasing on microcrystals | | Descriptor: | CALCIUM ION, Concanavalin-A,Concanavalin-A, MANGANESE (II) ION | | Authors: | Cianci, M, Nanao, M, Schneider, T.R. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Long-wavelength Mesh&Collect native SAD phasing from microcrystals.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2WR6
 
 | | Structure of the complex of RBP4 with linoleic acid | | Descriptor: | (11E,13E,15Z)-OCTADECA-11,13,15-TRIENOIC ACID, CHLORIDE ION, RETINOL-BINDING PROTEIN 4 | | Authors: | Huang, H.-J, Nanao, M, Stout, T, Rosen, J. | | Deposit date: | 2009-08-29 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a Non-Retinoid Compound and Fatty Acids as Ligands for Retinol Binding Protein 4 and Their Implications in Diabetes
To be Published
|
|
2BMF
 
 | | Dengue virus RNA helicase at 2.4A | | Descriptor: | RNA HELICASE | | Authors: | Xu, T, Sampath, A, Chao, A, Wen, D, Nanao, M, Chene, P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2005-03-14 | | Release date: | 2005-08-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of the Dengue Virus Helicase/Nucleoside Triphosphatase Catalytic Domain at a Resolution of 2.4 A.
J.Virol., 79, 2005
|
|
2BHR
 
 | | Dengue virus RNA helicase | | Descriptor: | RNA HELICASE, SULFATE ION | | Authors: | Xu, T, Sampath, A, Chao, A, Wen, D, Nanao, M, Chene, P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2005-01-17 | | Release date: | 2005-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Dengue Virus Helicase/Nucleoside Triphosphatase Catalytic Domain at a Resolution of 2.4 A.
J.Virol., 79, 2005
|
|
7QGF
 
 | | Cubic Insulin SAD phasing at 14.2 keV | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Nanao, M.H, Basu, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.203 Å) | | Cite: | ID23-2: an automated and high-performance microfocus beamline for macromolecular crystallography at the ESRF.
J.Synchrotron Radiat., 29, 2022
|
|
7PKU
 
 | | Structure of SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a | | Descriptor: | 3C-like proteinase, Nucleoprotein | | Authors: | Bessa, L.M, Guseva, S, Camacho-Zarco, A.R, Salvi, N, Blackledge, M. | | Deposit date: | 2021-08-26 | | Release date: | 2022-01-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a.
Sci Adv, 8, 2022
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
4NJ7
 
 | | PB1 Domain of AtARF7 - SeMet Derivative | | Descriptor: | Auxin response factor 7 | | Authors: | Korasick, D.A, Westfall, C.S, Lee, S.G, Nanao, M, Jez, J.M, Strader, L.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-03-26 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NJ6
 
 | | PB1 Domain of AtARF7 | | Descriptor: | Auxin response factor 7 | | Authors: | Korasick, D.A, Westfall, C.S, Lee, S.G, Nanao, M, Jez, J.M, Strader, L.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4WSB
 
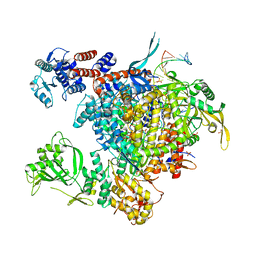 | | Bat Influenza A polymerase with bound vRNA promoter | | Descriptor: | Influenza A polymerase vRNA promoter 3' end, Influenza A polymerase vRNA promoter 5' end, PHOSPHATE ION, ... | | Authors: | Cusack, S, Pflug, A, Guilligay, D, Reich, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
4WRT
 
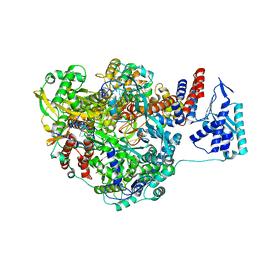 | | Crystal structure of Influenza B polymerase with bound vRNA promoter (form FluB2) | | Descriptor: | Influenza virus polymerase vRNA promoter 3' end, Influenza virus polymerase vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-25 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
4WSA
 
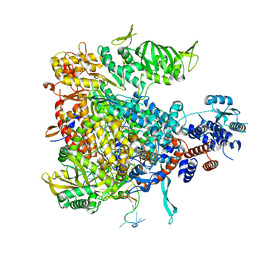 | | Crystal structure of Influenza B polymerase bound to the vRNA promoter (FluB1 form) | | Descriptor: | Influenza B vRNA promoter 3' end, Influenza B vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
4OX0
 
 | |
5MLM
 
 | | Plantago Major multifunctional oxidoreductase V150M mutant in complex with progesterone and NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, Progesterone 5-beta-reductase | | Authors: | Fellows, R, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2016-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.563 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
5MLH
 
 | | Plantago Major multifunctional oxidoreductase in complex with 8-oxogeranial and NADP+ | | Descriptor: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, CALCIUM ION, GLYCEROL, ... | | Authors: | Fellows, R, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2016-12-06 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
5MLR
 
 | | Plantago Major multifunctional oxidoreductase V150M mutant in complex with citral and NADP+ | | Descriptor: | CHLORIDE ION, Geranaldehyde, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Fellows, R, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2016-12-07 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
6F7J
 
 | |
6F7S
 
 | |
6F7P
 
 | |
6F8D
 
 | |
6GSD
 
 | | Plantago Major multifunctional oxidoreductase in complex with progesterone and NADP+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROGESTERONE, Progesterone 5-beta-reductase | | Authors: | Fellows, R, Silva, C, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
