4K17
 
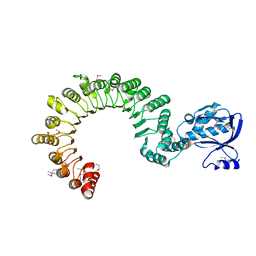 | | Crystal Structure of mouse CARMIL residues 1-668 | | Descriptor: | CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, Leucine-rich repeat-containing protein 16A, ... | | Authors: | Zwolak, A, Dominguez, R. | | Deposit date: | 2013-04-04 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | CARMIL leading edge localization depends on a non-canonical PH domain and dimerization.
Nat Commun, 4, 2013
|
|
2KZ7
 
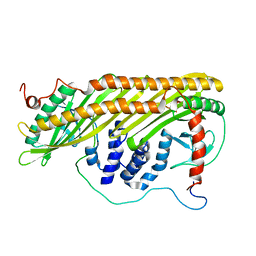 | |
2KXP
 
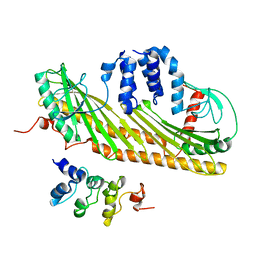 | |
5V7X
 
 | | Crystal Structure of Myosin 1b residues 1-728 with bound sulfate and Calmodulin | | Descriptor: | Calmodulin-1, SULFATE ION, Unconventional myosin-Ib | | Authors: | Zwolak, A, Shuman, H, Dominguez, R, Ostap, E.M. | | Deposit date: | 2017-03-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.141 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5UBX
 
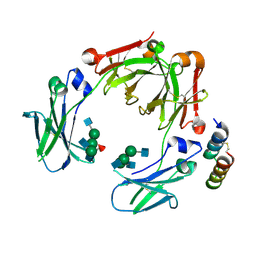 | | Crystal structure of a mutant mIgG2b Fc heterodimer in complex with Protein A peptide analog Z34C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-2B chain C region, ... | | Authors: | Armstrong, A.A, Zwolak, A, Gilliland, G.L. | | Deposit date: | 2016-12-21 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modulation of protein A binding allows single-step purification of mouse bispecific antibodies that retain FcRn binding.
MAbs, 9, 2017
|
|
6C1H
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C1D
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C1G
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4L79
 
 | | Crystal Structure of nucleotide-free Myosin 1b residues 1-728 with bound Calmodulin | | Descriptor: | Calmodulin, MAGNESIUM ION, Unconventional myosin-Ib | | Authors: | Shuman, H, Zwolak, A, Dominguez, R, Ostap, E.M. | | Deposit date: | 2013-06-13 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A vertebrate myosin-I structure reveals unique insights into myosin mechanochemical tuning.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2L5A
 
 | | Structural basis for recognition of centromere specific histone H3 variant by nonhistone Scm3 | | Descriptor: | Histone H3-like centromeric protein CSE4, Protein SCM3, Histone H4 | | Authors: | Zhou, Z, Feng, H, Zhou, B, Ghirlando, R, Hu, K, Zwolak, A, Jenkins, L, Xiao, H, Tjandra, N, Wu, C, Bai, Y. | | Deposit date: | 2010-10-28 | | Release date: | 2011-03-16 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.
Nature, 472, 2011
|
|
7T86
 
 | | Crystal Structure of Fab CR5133 / Phospho-SD Peptide Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab CR5133 Heavy Chain, Antibody Fab CR5133 Light Chain, ... | | Authors: | Luo, J, Malia, T.J, Buckley, P.T. | | Deposit date: | 2021-12-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
7T87
 
 | | CRYSTAL STRUCTURE OF LEUKOCIDIN AB/CENTYRIN S17/FAB 214F COMPLEX | | Descriptor: | Antibody Fab B214 Heavy Chain, Antibody Fab B214 Light Chain, Centyrin S17, ... | | Authors: | Luo, J, Malia, T.J, Buckley, P.T. | | Deposit date: | 2021-12-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
7T82
 
 | | Crystal Structure of LEUKOCIDIN E/CENTYRIN S26/FAB B438 | | Descriptor: | Antibody Fab Heavy Chain, Antibody Fab Light Chain, Centyrin S26, ... | | Authors: | Luo, J, Malia, T.J, Buckley, P.T. | | Deposit date: | 2021-12-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
8DY5
 
 | |
8DY0
 
 | | Crystal Structure of spFv GLK1 HL | | Descriptor: | spFv GLK1 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY1
 
 | |
8DY4
 
 | | Crystal Structure of spFv CAT2200 HL | | Descriptor: | spFv CAT2200 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY3
 
 | | Crystal Structure of spFv GLK2 HL | | Descriptor: | SULFATE ION, spFv GLK2 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY2
 
 | | Crystal Structure of spFv GLK1 | | Descriptor: | SULFATE ION, spFv GLK1 LH | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
