4MZM
 
 | | MazF from S. aureus crystal form I, P212121, 2.1 A | | Descriptor: | mRNA interferase MazF | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
4MZP
 
 | | MazF from S. aureus crystal form III, C2221, 2.7 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
4MZT
 
 | | MazF from S. aureus crystal form II, C2221, 2.3 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
5DLO
 
 | | S. aureus MazF in complex with substrate analogue | | Descriptor: | CHLORIDE ION, DNA substrate analogue AUACAUA, Endoribonuclease MazF | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-09-07 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Substrate recognition, regulation mechanism and activity regulation of MazF mRNA interferase.
To Be Published
|
|
2MF2
 
 | |
2MRN
 
 | |
2MRU
 
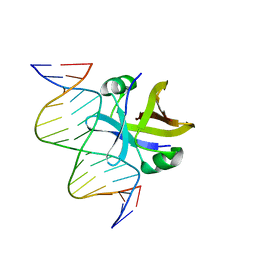 | | Structure of truncated EcMazE-DNA complex | | Descriptor: | Antitoxin MazE, DNA (5'-D(*CP*GP*TP*GP*AP*TP*AP*TP*AP*TP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*CP*TP*AP*TP*AP*TP*AP*TP*CP*AP*CP*G)-3') | | Authors: | Zorzini, V, Buts, L, Loris, R, van Nuland, N. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Escherichia coli antitoxin MazE as transcription factor: insights into MazE-DNA binding.
Nucleic Acids Res., 43, 2015
|
|
5CKF
 
 | | E. coli MazF E24A form I | | Descriptor: | Endoribonuclease MazF, SODIUM ION | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CKB
 
 | | E. coli MazF form II | | Descriptor: | Endoribonuclease MazF | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CKE
 
 | | E.coli MazF E24A form IIa | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Endoribonuclease MazF, SULFATE ION | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CKD
 
 | | E. coli MazF E24A form III | | Descriptor: | Endoribonuclease MazF, SODIUM ION | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CO7
 
 | | E. coli MazF form III | | Descriptor: | Endoribonuclease MazF | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-19 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.489 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CK9
 
 | | E. coli MazF form I | | Descriptor: | Endoribonuclease MazF, PENTAETHYLENE GLYCOL | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CKH
 
 | | E. coli MazF E24A form IIb | | Descriptor: | Endoribonuclease MazF | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
5CQX
 
 | |
5CR2
 
 | |
5CQY
 
 | |
6Z72
 
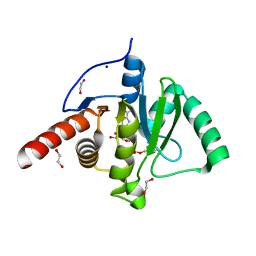 | | SARS-CoV-2 Macrodomain in complex with ADP-HPM | | Descriptor: | 1,2-ETHANEDIOL, Adenosine Diphosphate (Hydroxymethyl)pyrrolidine monoalcohol, D-MALATE, ... | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
6Z5T
 
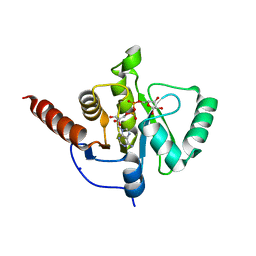 | | SARS-CoV-2 Macrodomain in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Replicase polyprotein 1ab, SODIUM ION | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-27 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
6Z6I
 
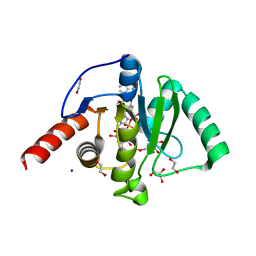 | | SARS-CoV-2 Macrodomain in complex with ADP-HPD | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, ... | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
7AQM
 
 | | ADP-ribosylserine hydrolase ARH3 of Latimeria chalumnae in complex with alpha-1''-O-methyl-ADP-ribose (meADPr) | | Descriptor: | ADP-ribosylhydrolase like 2, Adenosine 5'-diphosphoric acid beta-[(3beta,4beta-dihydroxy-5beta-methoxytetrahydrofuran-2alpha-yl)methyl] estere, MAGNESIUM ION | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
7ARW
 
 | | Structure of human ARH3 E41A bound to alpha-NAD+ and magnesium | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADP-ribose glycohydrolase ARH3, ... | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
7AKR
 
 | |
7AKS
 
 | |
6HHX
 
 | | Structure of T. thermophilus AspRS in Complex with 5'-O-(N-(L-aspartyl)-sulfamoyl)cytidine | | Descriptor: | 5'-O-(N-(L-aspartyl)-sulfamoyl)cytidine, Aspartate--tRNA(Asp) ligase | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
