2F68
 
 | |
2F6A
 
 | |
3V64
 
 | | Crystal Structure of agrin and LRP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low-density lipoprotein receptor-related protein 4, ... | | Authors: | Zong, Y, Zhang, B, Gu, S, Lee, K, Zhou, J, Yao, G, Figueiedo, D, Perry, K, Mei, L, Jin, R. | | Deposit date: | 2011-12-18 | | Release date: | 2012-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of agrin-LRP4-MuSK signaling.
Genes Dev., 26, 2012
|
|
3V65
 
 | | Crystal structure of agrin and LRP4 complex | | Descriptor: | Agrin, CALCIUM ION, Low-density lipoprotein receptor-related protein 4 | | Authors: | Zong, Y, Jin, R. | | Deposit date: | 2011-12-18 | | Release date: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of agrin-LRP4-MuSK signaling.
Genes Dev., 26, 2012
|
|
1QWZ
 
 | | Crystal structure of Sortase B from S. aureus complexed with MTSET | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, NICKEL (II) ION, NPQTN specific sortase B, ... | | Authors: | Zong, Y, Mazmanian, S.K, Schneewind, O, Narayana, S.V. | | Deposit date: | 2003-09-03 | | Release date: | 2004-04-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall
Structure, 12, 2004
|
|
1QX6
 
 | | Crystal structure of Sortase B complexed with E-64 | | Descriptor: | N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, NPQTN specific sortase B | | Authors: | Zong, Y, Mazmanian, S.K, Schneewind, O, Narayana, S.V. | | Deposit date: | 2003-09-04 | | Release date: | 2004-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall
Structure, 12, 2004
|
|
1QXA
 
 | | Crystal structure of Sortase B complexed with Gly3 | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, NPQTN specific sortase B, peptide GLY-GLY-GLY | | Authors: | Zong, Y, Mazmanian, S.K, Schneewind, O, Narayana, S.V. | | Deposit date: | 2003-09-05 | | Release date: | 2004-04-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall
Structure, 12, 2004
|
|
1T2W
 
 | | Crystal Structure of Sortase A in Complex with a LPETG peptide | | Descriptor: | Class A sortase SrtA, Peptide LEU-PRO-GLU-THR-GLY | | Authors: | Zong, Y, Bice, T.W, Ton-That, H, Schneewind, O, Narayana, S.V. | | Deposit date: | 2004-04-23 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Staphylococcus aureus sortase A and its substrate complex
J.Biol.Chem., 279, 2004
|
|
1T2O
 
 | | Crystal structure of Se-SrtA, C184-Ala | | Descriptor: | sortase | | Authors: | Zong, Y, Bice, T.W, Ton-That, H, Schneewind, O, Narayana, S.V. | | Deposit date: | 2004-04-22 | | Release date: | 2004-09-07 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Staphylococcus aureus sortase A and its substrate complex
J.Biol.Chem., 279, 2004
|
|
1T2P
 
 | | Crystal structure of Sortase A from Staphylococcus aureus | | Descriptor: | sortase | | Authors: | Zong, Y, Bice, T.W, Ton-That, H, Schneewind, O, Narayana, S.V. | | Deposit date: | 2004-04-22 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURES OF STAPHYLOCOCCUS AUREUS SORTASE A AND ITS SUBSTRATE COMPLEX
J.Biol.Chem., 279, 2004
|
|
3OBR
 
 | |
5E85
 
 | | isolated SBD of BiP | | Descriptor: | 78 kDa glucose-regulated protein, SULFATE ION | | Authors: | Liu, Q, Yang, J, Nune, M, Zong, Y, Zhou, L. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP.
Structure, 23, 2015
|
|
5E86
 
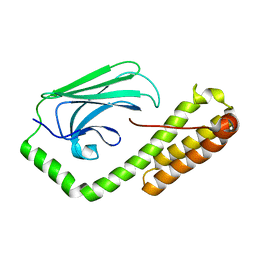 | | isolated SBD of BiP with loop34 modification | | Descriptor: | 78 kDa glucose-regulated protein | | Authors: | Liu, Q, Yang, J, Nune, M, Zong, Y, Zhou, L. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP.
Structure, 23, 2015
|
|
5E84
 
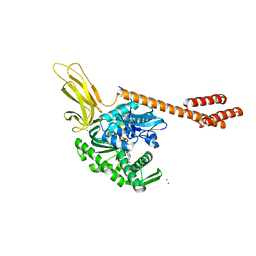 | | ATP-bound state of BiP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, Q, Yang, J, Nune, M, Zong, Y, Zhou, L. | | Deposit date: | 2015-10-13 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP.
Structure, 23, 2015
|
|
3OBT
 
 | |
3SAJ
 
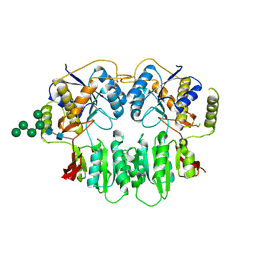 | | Crystal Structure of glutamate receptor GluA1 Amino Terminal Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 1, ... | | Authors: | Jin, R, Zong, Y, Yao, G, Gu, S. | | Deposit date: | 2011-06-02 | | Release date: | 2011-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the glutamate receptor GluA1 N-terminal domain.
Biochem.J., 438, 2011
|
|
6ASY
 
 | | BiP-ATP2 | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Liu, Q, Yang, J, Zong, Y, Columbus, L, Zhou, L. | | Deposit date: | 2017-08-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s.
Nat Commun, 8, 2017
|
|
7WB2
 
 | | Oxidase ChaP-D49L/Y109F mutant | | Descriptor: | ChaP, FE (III) ION | | Authors: | Zong, Y, Zheng, W, Wang, Y, Zhu, J, Tan, R. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alteration of the Catalytic Reaction Trajectory of a Vicinal Oxygen Chelate Enzyme by Directed Evolution.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
3QNQ
 
 | | Crystal structure of the transporter ChbC, the IIC component from the N,N'-diacetylchitobiose-specific phosphotransferase system | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, PTS system, ... | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Crystal structure of a phosphorylation-coupled saccharide transporter.
Nature, 473, 2011
|
|
7F04
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with Heme and ATP. | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F03
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F02
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7XHQ
 
 | | Small-molecule Allosteric Regulation Mechanism of SHP2 | | Descriptor: | 2-[(3S,4R)-4-azanyl-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-5-[2,3-bis(chloranyl)phenyl]-3-methyl-pyrrolo[2,1-f][1,2,4]triazin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, Y, Zhu, J, Yu, K, Liu, B. | | Deposit date: | 2022-04-09 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the SHP2 allosteric inhibitor 2-((3R,4R)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl)-5-(2,3-dichlorophenyl)-3-methylpyrrolo[2,1-f][1,2,4] triazin-4(3H)-one.
J Enzyme Inhib Med Chem, 38, 2023
|
|
7VFP
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with heme and single ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7VFJ
 
 | | Cytochrome c-type biogenesis protein CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Zhu, J.P, Zhang, K, Li, J, Zheng, W, Gu, M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
