2V03
 
 | | High resolution structure and catalysis of an O-acetylserine sulfhydrylase | | Descriptor: | CITRIC ACID, Cysteine synthase B, GLYCEROL, ... | | Authors: | Zocher, G, Wiesand, U, Schulz, G.E. | | Deposit date: | 2007-05-08 | | Release date: | 2007-10-09 | | Last modified: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High resolution structure and catalysis of O-acetylserine sulfhydrylase isozyme B from Escherichia coli.
FEBS J., 274, 2007
|
|
4YUF
 
 | |
4YUC
 
 | | Crystal Structure of CorB derivatized with S-(2-acetamidoethyl) 4-methyl-3-oxohexanethioate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CorB, SODIUM ION | | Authors: | Zocher, G, Vilstrup, J, Stehle, T. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-30 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural basis of head to head polyketide fusion by CorB.
Chem Sci, 6, 2015
|
|
5C1J
 
 | |
4EE6
 
 | |
4EE7
 
 | |
4EE8
 
 | |
4Q4W
 
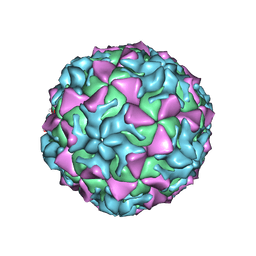 | |
4Q4V
 
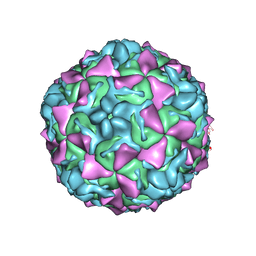 | | Crystal structure of Coxsackievirus A24v | | Descriptor: | Coxsackievirus capsid protein VP1, Coxsackievirus capsid protein VP2, Coxsackievirus capsid protein VP3, ... | | Authors: | Zocher, G, Stehle, T. | | Deposit date: | 2014-04-15 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A sialic Acid binding site in a human picornavirus.
Plos Pathog., 10, 2014
|
|
4Q4X
 
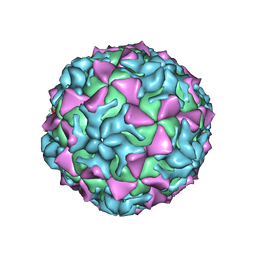 | |
4Q4Y
 
 | |
3P3O
 
 | | Crystal Structure of the Cytochrome P450 Monooxygenase AurH (ntermII) from Streptomyces Thioluteus | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zocher, G, Richter, M.E.A, Mueller, U, Hertweck, C. | | Deposit date: | 2010-10-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural fine-tuning of a multifunctional cytochrome p450 monooxygenase.
J.Am.Chem.Soc., 133, 2011
|
|
3P3X
 
 | | Crystal Structure of the Cytochrome P450 Monooxygenase AurH (nterm-AurH-I) from Streptomyces Thioluteus | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zocher, G, Richter, M.E.A, Mueller, U, Hertweck, C. | | Deposit date: | 2010-10-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural fine-tuning of a multifunctional cytochrome p450 monooxygenase.
J.Am.Chem.Soc., 133, 2011
|
|
3P3Z
 
 | | Crystal Structure of the Cytochrome P450 Monooxygenase AurH from Streptomyces Thioluteus in Complex with Ancymidol | | Descriptor: | (S)-cyclopropyl(4-methoxyphenyl)pyrimidin-5-ylmethanol, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zocher, G, Richter, M.E.A, Mueller, U, Hertweck, C. | | Deposit date: | 2010-10-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural fine-tuning of a multifunctional cytochrome p450 monooxygenase.
J.Am.Chem.Soc., 133, 2011
|
|
3P3L
 
 | | Crystal Structure of the Cytochrome P450 monooxygenase AurH (wildtype) from Streptomyces Thioluteus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zocher, G, Richter, M.E.A, Mueller, U, Hertweck, C. | | Deposit date: | 2010-10-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural fine-tuning of a multifunctional cytochrome p450 monooxygenase.
J.Am.Chem.Soc., 133, 2011
|
|
7QB5
 
 | |
6TSD
 
 | |
7OJ7
 
 | |
2WBS
 
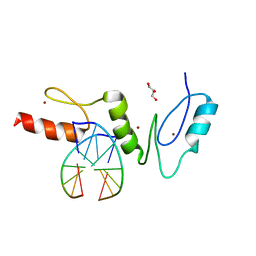 | | Crystal structure of the zinc finger domain of Klf4 bound to its target DNA | | Descriptor: | 5'-D(*GP*AP*GP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP*CP*TP*CP)-3', GLYCEROL, ... | | Authors: | Zocher, G, Schuetz, A, Carstanjen, D, Heinemann, U. | | Deposit date: | 2009-03-03 | | Release date: | 2010-04-07 | | Last modified: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation.
Cell.Mol.Life Sci., 68, 2011
|
|
4KC5
 
 | |
4LD7
 
 | | Crystal structure of AnaPT from Neosartorya fischeri | | Descriptor: | Dimethylallyl tryptophan synthase, SODIUM ION, TRIHYDROGEN THIODIPHOSPHATE | | Authors: | Zocher, G, Stehle, T. | | Deposit date: | 2013-06-24 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Catalytic Mechanism of Stereospecific Formation of cis-Configured Prenylated Pyrroloindoline Diketopiperazines by Indole Prenyltransferases.
Chem.Biol., 20, 2013
|
|
3T5Y
 
 | | Crystal structure of CerJ from Streptomyces tendae - malonic acid covalently linked to the catalytic Cystein C116 | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3T8E
 
 | | Crystal structure of CerJ from Streptomyces tendae soaked with CerviK | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-08-01 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3T6S
 
 | | Crystal structure of CerJ from Streptomyces tendae in Complex with CoA | | Descriptor: | COENZYME A, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-29 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
7A9C
 
 | |
