3DIN
 
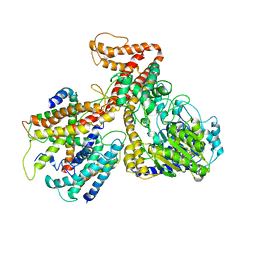 | | Crystal structure of the protein-translocation complex formed by the SecY channel and the SecA ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Zimmer, J, Nam, Y, Rapoport, T.A. | | Deposit date: | 2008-06-20 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a complex of the ATPase SecA and the protein-translocation channel.
Nature, 455, 2008
|
|
3JUX
 
 | |
3JV2
 
 | | Crystal Structure of B. subtilis SecA with bound peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein translocase subunit SecA, ... | | Authors: | Zimmer, J. | | Deposit date: | 2009-09-15 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational flexibility and peptide interaction of the translocation ATPase SecA.
J.Mol.Biol., 394, 2009
|
|
2IBM
 
 | |
4HG6
 
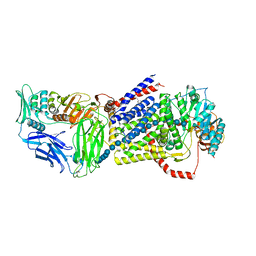 | |
6WLB
 
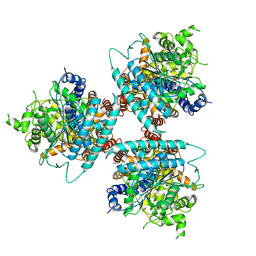 | | Structure of homotrimeric poplar cellulose synthase isoform 8 | | Descriptor: | Cellulose synthase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Zimmer, J, Pallinti, P, Ho, R. | | Deposit date: | 2020-04-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Architecture of a catalytically active homotrimeric plant cellulose synthase complex.
Science, 369, 2020
|
|
3DL8
 
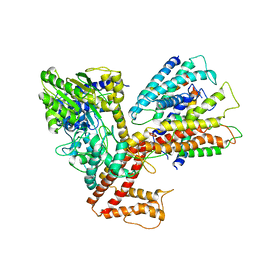 | | Structure of the complex of aquifex aeolicus SecYEG and bacillus subtilis SecA | | Descriptor: | Preprotein translocase subunit secY, Protein translocase subunit secA, Protein-export membrane protein secG, ... | | Authors: | Nam, Y, Zimmer, J, Rapoport, T.A. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (7.5 Å) | | Cite: | Structure of a complex of the ATPase SecA and the protein-translocation channel.
Nature, 455, 2008
|
|
6AN5
 
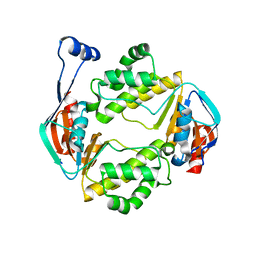 | |
6AMX
 
 | | Crystal Structure of Nucelotide Binding Domain of O-antigen polysaccharide ABC-transporter | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ABC transporter | | Authors: | Zimmer, J, Bi, Y. | | Deposit date: | 2017-08-11 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Architecture of a channel-forming O-antigen polysaccharide ABC transporter.
Nature, 553, 2018
|
|
3QXF
 
 | |
3QXQ
 
 | | Structure of the bacterial cellulose synthase subunit Z in complex with cellopentaose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Zimmer, J. | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Apo- and cellopentaose-bound structures of the bacterial cellulose synthase subunit BcsZ.
J.Biol.Chem., 286, 2011
|
|
8TSI
 
 | |
8TSH
 
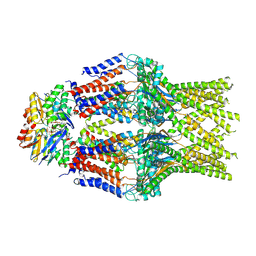 | |
8TT3
 
 | | S. thermodepolymerans KpsM-KpsE in Glycolipid 2 state with rigid body fitted KpsT | | Descriptor: | (2R,5S,8S)-2,5-dihydroxy-5,10-dioxo-8-[(undecanoyloxy)methyl]-4,6,9-trioxa-5lambda~5~-phosphahenicosan-1-yl 3-deoxy-alpha-L-altro-oct-2-ulopyranosidonic acid, ABC transporter ATP-binding protein, Capsular biosynthesis protein, ... | | Authors: | Kuklewicz, J, Zimmer, J. | | Deposit date: | 2023-08-12 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular insights into capsular polysaccharide secretion.
Nature, 2024
|
|
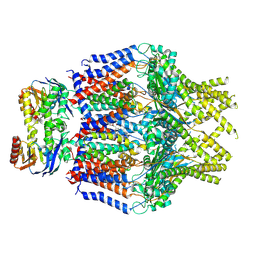
8TSW
 
 | | S. thermodepolymerans KpsMT-KpsE Apo 1 | | Descriptor: | ABC transporter ATP-binding protein, Capsular biosynthesis protein, Transport permease protein | | Authors: | Kuklewicz, J, Zimmer, J. | | Deposit date: | 2023-08-12 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into capsular polysaccharide secretion.
Nature, 2024
|
|
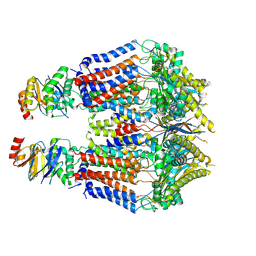
8TUN
 
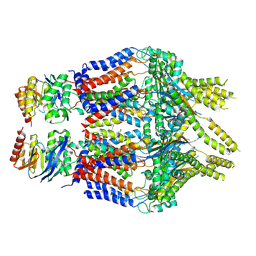 | | S. thermodepolymerans KpsM-KpsE in Glycolipid 1 state with rigid body fitted KpsT | | Descriptor: | (2R,5S,8S)-2,5-dihydroxy-5,10-dioxo-8-[(undecanoyloxy)methyl]-4,6,9-trioxa-5lambda~5~-phosphahenicosan-1-yl 3-deoxy-alpha-L-altro-oct-2-ulopyranosidonic acid, ABC transporter ATP-binding protein, Capsular biosynthesis protein, ... | | Authors: | Kuklewicz, J, Zimmer, J. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular insights into capsular polysaccharide secretion.
Nature, 2024
|
|
8TSL
 
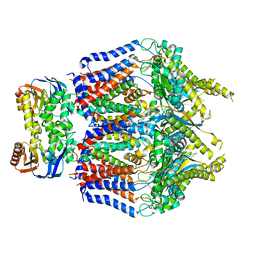 | |
5L22
 
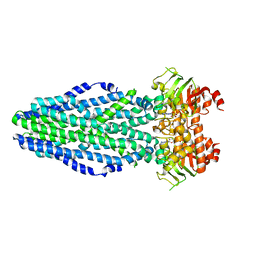 | | PrtD T1SS ABC transporter | | Descriptor: | ABC transporter (HlyB subfamily), ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Morgan, J.L.W, Zimmer, J. | | Deposit date: | 2016-07-30 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of a Type-1 Secretion System ABC Transporter.
Structure, 25, 2017
|
|
6O14
 
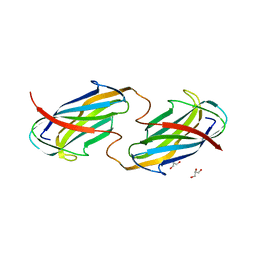 | |
8G2J
 
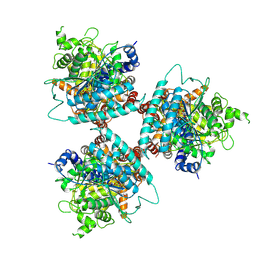 | |
8G27
 
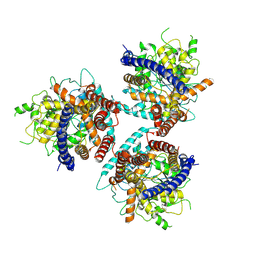 | | Hybrid aspen cellulose synthase-8 bound to UDP | | Descriptor: | Cellulose synthase, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Verma, P, Zimmer, J. | | Deposit date: | 2023-02-03 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Insights into substrate coordination and glycosyl transfer of poplar cellulose synthase-8.
Biorxiv, 2023
|
|
6M96
 
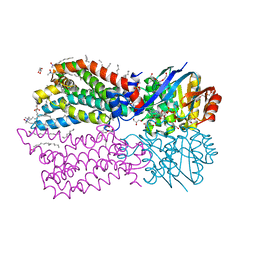 | |
5EIY
 
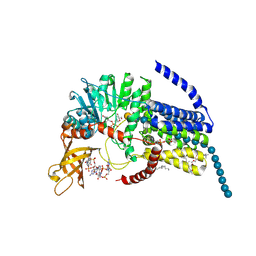 | | Bacterial cellulose synthase bound to a substrate analogue | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIUNDECYL PHOSPHATIDYL CHOLINE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | McNamara, J.T, Zimmer, J. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Observing cellulose biosynthesis and membrane translocation in crystallo.
Nature, 531, 2016
|
|
5EJZ
 
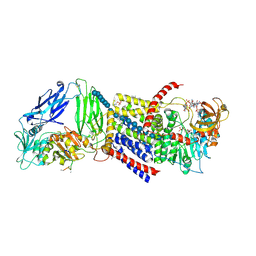 | | Bacterial Cellulose Synthase Product-Bound State | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-deoxy-2-fluoro-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ... | | Authors: | Morgan, J.L.W, Zimmer, J. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Observing cellulose biosynthesis and membrane translocation in crystallo.
Nature, 531, 2016
|
|
7LBY
 
 | |
