1BZ9
 
 | |
1B0G
 
 | |
1KMI
 
 | | CRYSTAL STRUCTURE OF AN E.COLI CHEMOTAXIS PROTEIN, CHEZ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BICINE, Chemotaxis protein cheY, ... | | Authors: | Zhao, R, Collins, E.J, Bourret, R.B, Silversmith, R.E. | | Deposit date: | 2001-12-16 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ.
Nat.Struct.Biol., 9, 2002
|
|
1RHI
 
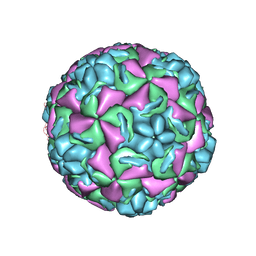 | | HUMAN RHINOVIRUS 3 COAT PROTEIN | | Descriptor: | CALCIUM ION, HUMAN RHINOVIRUS 3 COAT PROTEIN | | Authors: | Zhao, R, Rossmann, M.G. | | Deposit date: | 1996-06-17 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human rhinovirus 3 at 3.0 A resolution.
Structure, 4, 1996
|
|
1T5I
 
 | | Crystal structure of the C-terminal domain of UAP56 | | Descriptor: | C_TERMINAL DOMAIN OF A PROBABLE ATP-DEPENDENT RNA HELICASE | | Authors: | Zhao, R, Green, M.R, Shen, J. | | Deposit date: | 2004-05-04 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of UAP56, a "DEXD/H-box" protein involved in pre-mRNA splicing and mRNA export
Structure, 12, 2004
|
|
1T6N
 
 | | Crystal structure of the N-terminal domain of human UAP56 | | Descriptor: | CITRATE ANION, Probable ATP-dependent RNA helicase | | Authors: | Zhao, R. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of UAP56, a DExD/H-Box Protein Involved in Pre-mRNA Splicing and mRNA Export
Structure, 12, 2004
|
|
8JIX
 
 | |
6IYA
 
 | |
4EGC
 
 | |
2N6B
 
 | |
1AYN
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Oliveira, M.A, Zhao, R, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human rhinovirus 16.
Structure, 1, 1993
|
|
1D4Z
 
 | | CRYSTAL STRUCTURE OF CHEY-95IV, A HYPERACTIVE CHEY MUTANT | | Descriptor: | CHEMOTAXIS PROTEIN CHEY, SULFATE ION | | Authors: | Schuster, M, Zhao, R, Bourret, R.B, Collins, E.J. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Correlated switch binding and signaling in bacterial chemotaxis.
J.Biol.Chem., 275, 2000
|
|
1I7R
 
 | | CRYSTAL STRUCTURE OF CLASS I MHC A2 IN COMPLEX WITH PEPTIDE P1058 | | Descriptor: | 9 RESIDUE PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Busslep, J, Zhao, R, Loftus, D, Appella, E, Collins, E.J. | | Deposit date: | 2001-03-10 | | Release date: | 2001-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | T cell activity correlates with oligomeric peptide/MHC binding on T cell surface
J.Biol.Chem., 276, 2001
|
|
1I7U
 
 | | CRYSTAL STRUCTURE OF CLASS I MHC A2 IN COMPLEX WITH PEPTIDE P1049-6V | | Descriptor: | 9 RESIDUE PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Busslep, J, Zhao, R, Loftus, D, Appella, E, Collins, E.J. | | Deposit date: | 2001-03-10 | | Release date: | 2001-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | T cell activity correlates with oligomeric peptide-major histocompatibility complex binding on T cell surface
J.Biol.Chem., 276, 2001
|
|
1I7T
 
 | | CRYSTAL STRUCTURE OF CLASS I MHC A2 IN COMPLEX WITH PEPTIDE P1049-5V | | Descriptor: | 9 RESIDUE PEPTIDE, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Busslep, J, Zhao, R, Loftus, D, Appella, E, Collins, E.J. | | Deposit date: | 2001-03-10 | | Release date: | 2001-10-24 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | T cell activity correlates with oligomeric peptide-major histocompatibility complex binding on T cell surface
J.Biol.Chem., 276, 2001
|
|
1MIH
 
 | | A ROLE FOR CHEY GLU 89 IN CHEZ-MEDIATED DEPHOSPHORYLATION OF THE E. COLI CHEMOTAXIS RESPONSE REGULATOR CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, MANGANESE (II) ION, ... | | Authors: | Silversmith, R.E, Guanga, G.P, Betts, L, Chu, C, Zhao, R, Bourret, R.B. | | Deposit date: | 2002-08-23 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89.
J.Bacteriol., 185, 2003
|
|
5WWP
 
 | | Crystal structure of Middle East respiratory syndrome coronavirus helicase (MERS-CoV nsp13) | | Descriptor: | ORF1ab, SULFATE ION, ZINC ION | | Authors: | Hao, W, Wojdyla, J.A, Zhao, R, Han, R, Das, R, Zlatev, I, Manoharan, M, Wang, M, Cui, S. | | Deposit date: | 2017-01-03 | | Release date: | 2017-07-05 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Middle East respiratory syndrome coronavirus helicase
PLoS Pathog., 13, 2017
|
|
1N25
 
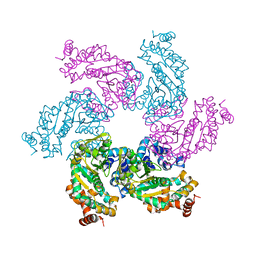 | | Crystal structure of the SV40 Large T antigen helicase domain | | Descriptor: | Large T Antigen, ZINC ION | | Authors: | Li, D, Zhao, R, Lilyestrom, W, Gai, D, Zhang, R, DeCaprio, J.A, Fanning, E, Jochimiak, A, Szakonyi, G, Chen, X.S. | | Deposit date: | 2002-10-21 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen
Nature, 423, 2003
|
|
1OOI
 
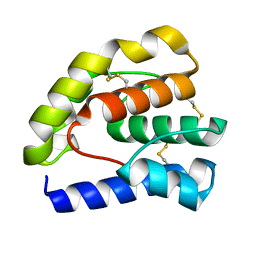 | |
1OOF
 
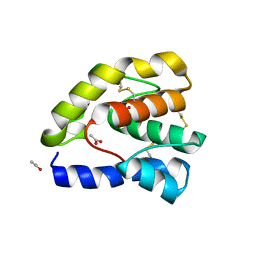 | | Complex of Drosophila odorant binding protein LUSH with ethanol | | Descriptor: | ACETATE ION, ETHANOL, odorant binding protein LUSH | | Authors: | Kruse, S.W, Zhao, R, Smith, D.P, Jones, D.N.M. | | Deposit date: | 2003-03-03 | | Release date: | 2003-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster
Nat.Struct.Biol., 10, 2003
|
|
1OOG
 
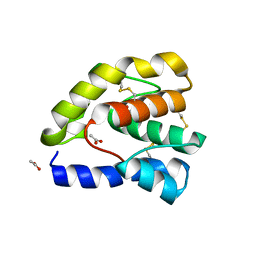 | | Complex of Drosophila odorant binding protein LUSH with propanol | | Descriptor: | ACETATE ION, N-PROPANOL, odorant binding protein LUSH | | Authors: | Kruse, S.W, Zhao, R, Smith, D.P, Jones, D.N.M. | | Deposit date: | 2003-03-03 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster
Nat.Struct.Biol., 10, 2003
|
|
1OOH
 
 | | Complex of Drosophila odorant binding protein LUSH with butanol | | Descriptor: | 1-BUTANOL, ACETATE ION, odorant binding protein LUSH | | Authors: | Kruse, S.W, Zhao, R, Smith, D.P, Jones, D.N.M. | | Deposit date: | 2003-03-03 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster
Nat.Struct.Biol., 10, 2003
|
|
1SVL
 
 | | Co-crystal structure of SV40 large T antigen helicase domain and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
1SVM
 
 | | Co-crystal structure of SV40 large T antigen helicase domain and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
1SVO
 
 | | Structure of SV40 large T antigen helicase domain | | Descriptor: | ZINC ION, large T antigen | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
