4N7R
 
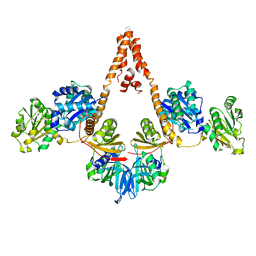 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its binding protein | | Descriptor: | Genomic DNA, chromosome 3, P1 clone: MXL8, ... | | Authors: | Zhao, A, Fang, Y, Lin, Y, Gong, W, Liu, L. | | Deposit date: | 2013-10-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5YJL
 
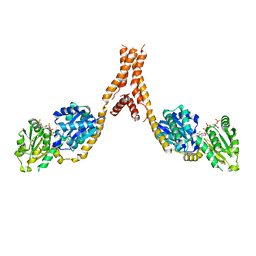 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with NADPH and GBP | | Descriptor: | Glutamyl-tRNA reductase 1, Glutamyl-tRNA reductase-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, A, Han, F. | | Deposit date: | 2017-10-11 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Arabidopsis thaliana glutamyl-tRNAGlureductase in complex with NADPH and glutamyl-tRNAGlureductase binding protein
Photosyn. Res., 137, 2018
|
|
3PPD
 
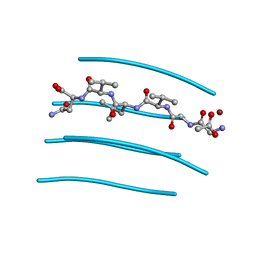 | | GGVLVN segment from Human Prostatic Acid Phosphatase Residues 260-265, involved in Semen-Derived Enhancer of Viral Infection | | Descriptor: | ACETIC ACID, GGVLVN peptide, amyloid forming segment, ... | | Authors: | Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation.
Nature, 475, 2011
|
|
5I3L
 
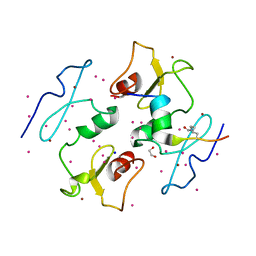 | | DPF3b in complex with H3K14ac peptide | | Descriptor: | 1,2-ETHANEDIOL, H3K14ac peptide, SODIUM ION, ... | | Authors: | Tempel, W, Liu, Y, Walker, J.R, Zhao, A, Qin, S, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-10 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of DPF3b in complex with an acetylated histone peptide.
J.Struct.Biol., 195, 2016
|
|
5IIW
 
 | | Corkscrew assembly of SOD1 residues 28-38 without potassium iodide | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Sangwan, S, Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS).
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5DLI
 
 | | Corkscrew assembly of SOD1 residues 28-38 | | Descriptor: | GLYCEROL, IODIDE ION, Superoxide dismutase [Cu-Zn] | | Authors: | Sangwan, S, Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2015-09-05 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS).
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4QDJ
 
 | |
4QDK
 
 | |
5CHE
 
 | |
4GG5
 
 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | 3-(4-methylpiperazin-1-yl)-N-(3-nitrobenzyl)-7-(trifluoromethyl)quinolin-5-amine, Hepatocyte growth factor receptor | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-08-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Multisubstituted quinoxalines and pyrido[2,3-d]pyrimidines: Synthesis and SAR study as tyrosine kinase c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4GG7
 
 | | Crystal structure of cMET in complex with novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, N-(3-nitrobenzyl)-6-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-2-(trifluoromethyl)pyrido[2,3-d]pyrimidin-4-amine | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Multisubstituted quinoxalines and pyrido[2,3-d]pyrimidines: Synthesis and SAR study as tyrosine kinase c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6E52
 
 | |
6E95
 
 | |
7F7W
 
 | | JAK2-JH2 | | Descriptor: | 2-((1-(2-fluoro-4-((4-(1-isopropyl-1H-pyrazol-4-yl)-5-methylpyrimidin-2-yl)amino)phenyl)piperidin-4-yl)(methyl)amino)ethan-1-ol, Tyrosine-protein kinase JAK2 | | Authors: | Niu, L. | | Deposit date: | 2021-06-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Preclinical studies of Flonoltinib Maleate, a novel JAK2/FLT3 inhibitor, in treatment of JAK2 V617F -induced myeloproliferative neoplasms.
Blood Cancer J, 12, 2022
|
|
5EOB
 
 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | 6-[bis(fluoranyl)-[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]quinoline, Hepatocyte growth factor receptor | | Authors: | Liu, Q, Chen, T, Xu, Y. | | Deposit date: | 2015-11-10 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of 6-(difluoro(6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl)methyl)quinoline as a highly potent and selective c-Met inhibitor
Eur.J.Med.Chem., 116, 2016
|
|
