4X6R
 
 | | An Isoform-specific Myristylation Switch Targets RIIb PKA Holoenzymes to Membranes | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Zhang, P, Ye, F, Bastidas, A.C, Kornev, A.P, Ginsberg, M.H, Wu, J, Taylor, S.S. | | Deposit date: | 2014-12-09 | | Release date: | 2015-07-22 | | Last modified: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Isoform-Specific Myristylation Switch Targets Type II PKA Holoenzymes to Membranes.
Structure, 23, 2015
|
|
4X8P
 
 | | Crystal structure of Ash2L SPRY domain in complex with RbBP5 | | Descriptor: | GLYCEROL, Retinoblastoma-binding protein 5, Set1/Ash2 histone methyltransferase complex subunit ASH2,Set1/Ash2 histone methyltransferase complex subunit ASH2 | | Authors: | Zhang, P, Chaturvedi, C.P, Brunzelle, J.S, Skiniotis, G, Brand, M, Shilatifard, A, Couture, J.-F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation.
Genes Dev., 29, 2015
|
|
4X6Q
 
 | | An Isoform-specific Myristylation Switch Targets RIIb PKA Holoenzymes to Membranes | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Zhang, P, Ye, F, Bastidas, A.C, Kornev, A.P, Ginsberg, M.H, Taylor, S.S. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | An Isoform-Specific Myristylation Switch Targets Type II PKA Holoenzymes to Membranes.
Structure, 23, 2015
|
|
4X8N
 
 | | Crystal structure of Ash2L SPRY domain in complex with phosphorylated RbBP5 | | Descriptor: | Retinoblastoma-binding protein 5, Set1/Ash2 histone methyltransferase complex subunit ASH2 | | Authors: | Zhang, P, Chaturvedi, C.P, Brunzelle, J.S, Skiniotis, G, Brand, M, Shilatifard, A, Couture, J.-F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation.
Genes Dev., 29, 2015
|
|
4WBB
 
 | | Single Turnover Autophosphorylation Cycle of the PKA RIIb Holoenzyme | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Zhang, P, Knape, M.J, Ahuja, L.G, Keshwani, M.M, King, C.C, Sastri, M, Herberg, F.W, Taylor, S.S. | | Deposit date: | 2014-09-02 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single Turnover Autophosphorylation Cycle of the PKA RII beta Holoenzyme.
Plos Biol., 13, 2015
|
|
1DRO
 
 | | NMR STRUCTURE OF THE CYTOSKELETON/SIGNAL TRANSDUCTION PROTEIN | | Descriptor: | BETA-SPECTRIN | | Authors: | Zhang, P, Talluri, S, Deng, H, Branton, D, Wagner, G. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the pleckstrin homology domain of Drosophila beta-spectrin.
Structure, 3, 1995
|
|
3F6G
 
 | | Crystal structure of the regulatory domain of LiCMS in complexed with isoleucine - type II | | Descriptor: | Alpha-isopropylmalate synthase, ISOLEUCINE, SULFATE ION, ... | | Authors: | Zhang, P, Ma, J, Zhao, G, Ding, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of the inhibitor selectivity and insights into the feedback inhibition mechanism of citramalate synthase from Leptospira interrogans
Biochem.J., 421, 2009
|
|
3F6H
 
 | | Crystal structure of the regulatory domain of LiCMS in complexed with isoleucine - type III | | Descriptor: | Alpha-isopropylmalate synthase, ISOLEUCINE, ZINC ION | | Authors: | Zhang, P, Ma, J, Zhao, G, Ding, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of the inhibitor selectivity and insights into the feedback inhibition mechanism of citramalate synthase from Leptospira interrogans
Biochem.J., 421, 2009
|
|
6VGU
 
 | |
3EPC
 
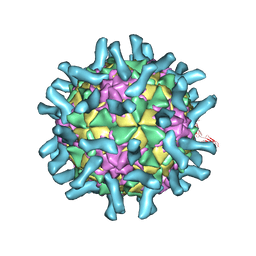 | | CryoEM structure of poliovirus receptor bound to poliovirus type 1 | | Descriptor: | MYRISTIC ACID, Poliovirus receptor, Protein VP1, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EPD
 
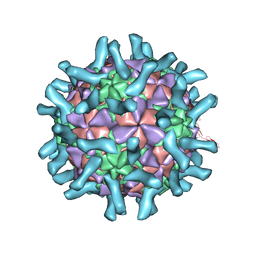 | | CryoEM structure of poliovirus receptor bound to poliovirus type 3 | | Descriptor: | MYRISTIC ACID, Poliovirus Type3 peptide, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EPF
 
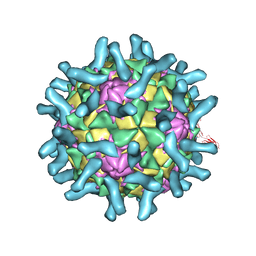 | | CryoEM structure of poliovirus receptor bound to poliovirus type 2 | | Descriptor: | 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE, MYRISTIC ACID, Poliovirus receptor, ... | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7FHA
 
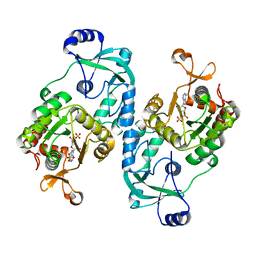 | | Crystal structure of the ATP sulfurylase domain of human PAPSS2 in complex with APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthase 2, POTASSIUM ION, ... | | Authors: | Zhang, P, Zhang, L, Zhang, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the substrate recognition mechanism of ATP-sulfurylase domain of human PAPS synthase 2.
Biochem.Biophys.Res.Commun., 586, 2022
|
|
7FH3
 
 | | Crystal structure of the ATP sulfurylase domain of human PAPSS2 | | Descriptor: | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthase 2, SULFATE ION, beta-D-glucopyranose | | Authors: | Zhang, P, Zhang, L, Zhang, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate recognition mechanism of ATP-sulfurylase domain of human PAPS synthase 2.
Biochem.Biophys.Res.Commun., 586, 2022
|
|
3TNQ
 
 | | Structure and Allostery of the PKA RIIb Tetrameric Holoenzyme | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein kinase, ... | | Authors: | Zhang, P, Smith-Nguyen, E.V, Keshwani, M.M, Deal, M.S, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-01 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structure and allostery of the PKA RIIbeta tetrameric holoenzyme
Science, 335, 2012
|
|
3TNP
 
 | | Structure and Allostery of the PKA RIIb Tetrameric Holoenzyme | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Zhang, P, Smith-Nguyen, E.V, Keshwani, M.M, Deal, M.S, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-01 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and allostery of the PKA RIIbeta tetrameric holoenzyme
Science, 335, 2012
|
|
3UVN
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of SET1A | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UVM
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL4 | | Descriptor: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3URO
 
 | | Poliovirus receptor CD155 D1D2 | | Descriptor: | Poliovirus receptor | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (3.5005 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3UVO
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of SET1B | | Descriptor: | Histone-lysine N-methyltransferase SETD1B, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UVL
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL3 | | Descriptor: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UVK
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase MLL2, SULFATE ION, ... | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
2F5J
 
 | | Crystal structure of MRG domain from human MRG15 | | Descriptor: | Mortality factor 4-like protein 1 | | Authors: | Zhang, P, Du, J, Ding, J. | | Deposit date: | 2005-11-26 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14
Protein Sci., 15, 2006
|
|
2F5K
 
 | |
3P5N
 
 | |
