4UFS
 
 | |
4UFR
 
 | |
3CJ9
 
 | |
3CJ7
 
 | | Structure of Rattus norvegicus NTPDase2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ectonucleoside triphosphate diphosphohydrolase 2 | | Authors: | Zebisch, M, Strater, N. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into signal conversion and inactivation by NTPDase2 in purinergic signaling
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CJ1
 
 | | Structure of Rattus norvegicus NTPDase2 | | Descriptor: | Ectonucleoside triphosphate diphosphohydrolase 2 | | Authors: | Zebisch, M, Strater, N. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into signal conversion and inactivation by NTPDase2 in purinergic signaling
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CJA
 
 | |
5FWT
 
 | | Wnt modulator Kremen crystal form I at 2.10A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
5FWU
 
 | | Wnt modulator Kremen crystal form II at 2.8A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, KREMEN PROTEIN 1 | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
5FWV
 
 | | Wnt modulator Kremen crystal form III at 3.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, KREMEN PROTEIN 1 | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
5FWS
 
 | | Wnt modulator Kremen crystal form I at 1.90A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
5FWW
 
 | | Wnt modulator Kremen in complex with DKK1 (CRD2) and LRP6 (PE3PE4) | | Descriptor: | CALCIUM ION, DICKKOPF-RELATED PROTEIN 1, KREMEN PROTEIN 1, ... | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
3ZX2
 
 | | NTPDase1 in complex with Decavanadate | | Descriptor: | ACETIC ACID, CHLORIDE ION, DECAVANADATE, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic Evidence for a Domain Motion in Rat Nucleoside Triphosphate Diphosphohydrolase (Ntpdase) 1.
J.Mol.Biol., 415, 2012
|
|
3ZX3
 
 | | Crystal Structure and Domain Rotation of NTPDase1 CD39 | | Descriptor: | ACETIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 1, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Evidence for a Domain Motion in Rat Nucleoside Triphosphate Diphosphohydrolase (Ntpdase) 1.
J.Mol.Biol., 415, 2012
|
|
3ZX0
 
 | | NTPDase1 in complex with Heptamolybdate | | Descriptor: | ACETIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 1, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic evidence for a domain motion in rat nucleoside triphosphate diphosphohydrolase (NTPDase) 1.
J. Mol. Biol., 415, 2012
|
|
4C8T
 
 | | Xenopus ZNRF3 ectodomain crystal form I | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4BRE
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with transition state mimic adenosine 5'phosphovanadate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-PHOSPHOVANADATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRN
 
 | | Legionella pneumophila NTPDase1 crystal form III (closed) in complex with Mg AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BR9
 
 | | Legionella pneumophila NTPDase1 crystal form II, closed, apo | | Descriptor: | ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, GLYCEROL, SULFATE ION | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRP
 
 | | Legionella pneumophila NTPDase1 crystal form V (part-open) | | Descriptor: | BROMIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRL
 
 | | Legionella pneumophila NTPDase1 crystal form III (closed) in complex with transition state mimic guanosine 5'-phosphovanadate | | Descriptor: | CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, GUANOSINE-5'-PHOSPHOVANADATE, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4C85
 
 | | zebrafish ZNRF3 ectodomain crystal form II | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-20 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C8C
 
 | | mouse ZNRF3 ectodomain crystal form III | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-20 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C8A
 
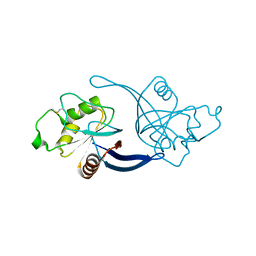 | | mouse ZNRF3 ectodomain crystal form II | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-20 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C8V
 
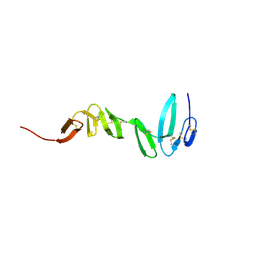 | | Xenopus RSPO2 Fu1-Fu2 crystal form I | | Descriptor: | R-SPONDIN-2 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C9E
 
 | |
