1VLK
 
 | |
2ILK
 
 | |
1ILK
 
 | |
1MFA
 
 | |
1IHT
 
 | |
1IHS
 
 | |
1MFB
 
 | |
1MFD
 
 | |
1MFC
 
 | |
3FIV
 
 | | CRYSTAL STRUCTURE OF FELINE IMMUNODEFICIENCY VIRUS PROTEASE COMPLEXED WITH A SUBSTRATE | | Descriptor: | ACE-ALN-VAL-LEU-ALA-GLU-ALN-NH2, FELINE IMMUNODEFICIENCY VIRUS PROTEASE, SULFATE ION | | Authors: | Schalk-Hihi, C, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1997-07-09 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor.
Biochemistry, 36, 1997
|
|
4FIV
 
 | | FIV PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, FELINE IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-15 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
2FIV
 
 | | Crystal structure of feline immunodeficiency virus protease complexed with a substrate | | Descriptor: | ACE-ALN-VAL-STA-GLU-ALN-NH2, FELINE IMMUNODEFICIENCY VIRUS PROTEASE, SULFATE ION | | Authors: | Schalk-Hihi, C, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1997-07-21 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor.
Biochemistry, 36, 1997
|
|
4CMS
 
 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES IV. STRUCTURE AND REFINEMENT AT 2.2 ANGSTROMS RESOLUTION OF BOVINE CHYMOSIN | | Descriptor: | CHYMOSIN B | | Authors: | Newman, M, Frazao, C, Khan, G, Tickle, I.J, Blundell, T.L, Safro, M, Andreeva, N, Zdanov, A. | | Deposit date: | 1991-11-01 | | Release date: | 1991-11-07 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray analyses of aspartic proteinases. IV. Structure and refinement at 2.2 A resolution of bovine chymosin.
J.Mol.Biol., 221, 1991
|
|
1ODY
 
 | | HIV-1 PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, HIV-1 PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
3OG4
 
 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P21212 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin 28 receptor, alpha (Interferon, ... | | Authors: | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
3OG6
 
 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Interleukin 28 receptor, ... | | Authors: | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
1ODX
 
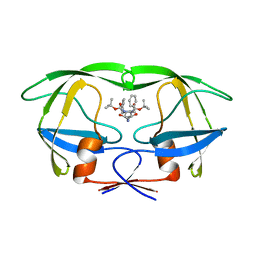 | | HIV-1 Proteinase mutant A71T, V82A | | Descriptor: | HIV-1 PROTEASE, di-tert-butyl {iminobis[(2S,3S)-3-hydroxy-1-phenylbutane-4,2-diyl]}biscarbamate | | Authors: | Kervinen, J, Thanki, N, Zdanov, A, Wlodawer, A. | | Deposit date: | 1996-09-16 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of the Native and Drug-Resistant HIV-1 Proteinases Complexed with an Aminodiol Inhibitor
Protein Pept.Lett., 3, 1996
|
|
1PV8
 
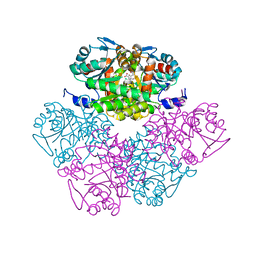 | | Crystal structure of a low activity F12L mutant of human porphobilinogen synthase | | Descriptor: | 3-(2-AMINOETHYL)-4-(AMINOMETHYL)HEPTANEDIOIC ACID, Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Breinig, S, Kervinen, J, Stith, L, Wasson, A.S, Fairman, R, Wlodawer, A, Zdanov, A, Jaffe, E.K. | | Deposit date: | 2003-06-26 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase.
Nat.Struct.Biol., 10, 2003
|
|
1QDM
 
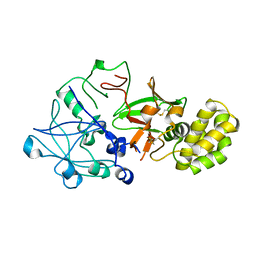 | | CRYSTAL STRUCTURE OF PROPHYTEPSIN, A ZYMOGEN OF A BARLEY VACUOLAR ASPARTIC PROTEINASE. | | Descriptor: | PROPHYTEPSIN | | Authors: | Kervinen, J, Tobin, G.J, Costa, J, Waugh, D.S, Wlodawer, A, Zdanov, A. | | Deposit date: | 1999-05-19 | | Release date: | 1999-07-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting.
EMBO J., 18, 1999
|
|
2FMB
 
 | | EIAV PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, EQUINE INFECTIOUS ANEMIA VIRUS PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
3GEF
 
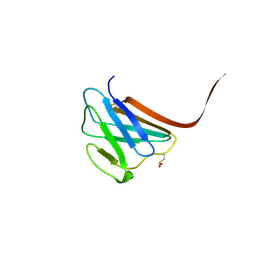 | | Crystal structure of the R482W mutant of lamin A/C | | Descriptor: | Lamin-A/C | | Authors: | Magracheva, E, Kozlov, S, Stuart, C, Wlodawer, A, Zdanov, A. | | Deposit date: | 2009-02-25 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the lamin A/C R482W mutant responsible for dominant familial partial lipodystrophy (FPLD).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2EMN
 
 | |
2EMD
 
 | |
2EMO
 
 | |
1FSI
 
 | | CRYSTAL STRUCTURE OF CYCLIC NUCLEOTIDE PHOSPHODIESTERASE OF APPR>P FROM ARABIDOPSIS THALIANA | | Descriptor: | CYCLIC PHOSPHODIESTERASE, SULFATE ION | | Authors: | Hofmann, A, Zdanov, A, Genschik, P, Filipowicz, W, Ruvinov, S, Wlodawer, A. | | Deposit date: | 2000-09-10 | | Release date: | 2000-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of activity of the cyclic phosphodiesterase of Appr>p, a product of the tRNA splicing reaction.
EMBO J., 19, 2000
|
|
