4XID
 
 | | AntpHD with 15bp DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2015-01-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
4X9J
 
 | | EGR-1 with Doubly Methylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*(5CM)P*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*(5CM)P*GP*C)-3'), Early growth response protein 1, ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2014-12-11 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1.
Febs Lett., 589, 2015
|
|
4XIC
 
 | | ANTPHD WITH 15BP di-thioate modified DNA DUPLEX | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C2S)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2015-01-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
5JLW
 
 | | AntpHD with 15bp DNA duplex R-monothioated at Cytidine-8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C7R)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J, Nguyen, D. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Stereospecific Effects of Oxygen-to-Sulfur Substitution in DNA Phosphate on Ion Pair Dynamics and Protein-DNA Affinity.
Chembiochem, 17, 2016
|
|
5JLX
 
 | | AntpHD with 15bp DNA duplex S-monothioated at Cytidine-8 | | Descriptor: | DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C7S)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), Homeotic protein antennapedia, ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J, Nguyen, D. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.748 Å) | | Cite: | Stereospecific Effects of Oxygen-to-Sulfur Substitution in DNA Phosphate on Ion Pair Dynamics and Protein-DNA Affinity.
Chembiochem, 17, 2016
|
|
6NTU
 
 | |
6VKQ
 
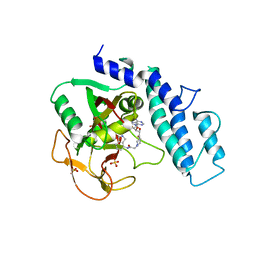 | | Crystal Structure of human PARP-1 CAT domain bound to inhibitor EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Steffen, J.D, Pascal, J.M. | | Deposit date: | 2020-01-21 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for allosteric PARP-1 retention on DNA breaks.
Science, 368, 2020
|
|
6VKK
 
 | |
6VKO
 
 | |
6BHV
 
 | | Human PARP-1 bound to NAD+ analog benzamide adenine dinucleotide (BAD) | | Descriptor: | Poly [ADP-ribose] polymerase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4R,5S)-5-(3-carbamoylphenyl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | Authors: | Pascal, J.M, Langelier, M.F. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NAD+analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains.
Nat Commun, 9, 2018
|
|
