5WT7
 
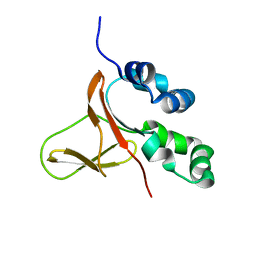 | | FAS1-IV domain of Human Periostin | | Descriptor: | Periostin | | Authors: | Yun, H, Lee, C.W. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | 1H,13C, and 15N resonance assignments of FAS1-IV domain of human periostin, a component of extracellular matrix proteins.
Biomol NMR Assign, 12, 2018
|
|
6M0Y
 
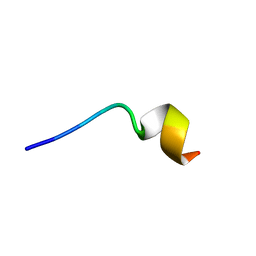 | | KR-12 analog derived from human LL-37 | | Descriptor: | LYS-ARG-ILE-VAL-LYS-ARG-ILE-LYS-LYS-TRP-LEU-ARG | | Authors: | Yun, H, Min, H.J, Lee, C.W. | | Deposit date: | 2020-02-24 | | Release date: | 2021-02-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Bactericidal Activity of KR-12 Analog Derived from Human LL-37 as a Potential Cosmetic Preservative
To Be Published
|
|
2MXD
 
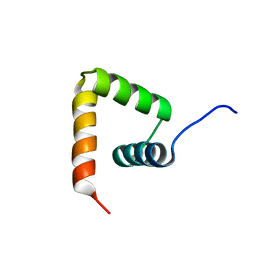 | | Solution structure of VPg of porcine sapovirus | | Descriptor: | Viral protein genome-linked | | Authors: | Kim, J, Hwang, H, Min, H, Yun, H, Cho, K, Pelton, J.G, Wemmer, D.E, Lee, C. | | Deposit date: | 2014-12-24 | | Release date: | 2015-04-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the porcine sapovirus VPg core reveals a stable three-helical bundle with a conserved surface patch.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
1YM4
 
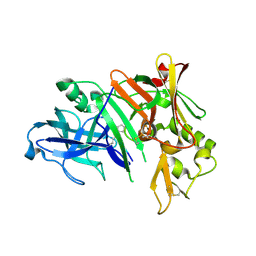 | | Crystal structure of human beta secretase complexed with NVP-AMK640 | | Descriptor: | Beta-secretase 1, NVP-AMK640 INHIBITOR | | Authors: | Hanessian, S, Yun, H, Hou, Y, Yang, G, Bayrakdarian, M, Therrien, E, Moitessier, N, Roggo, S, Veenstra, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
J.Med.Chem., 48, 2005
|
|
1YM2
 
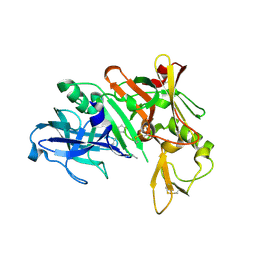 | | Crystal structure of human beta secretase complexed with NVP-AUR200 | | Descriptor: | Beta-secretase 1, NVP-AUR200 INHIBITOR | | Authors: | Hanessian, S, Yun, H, Hou, Y, Yang, G, Bayrakdarian, M, Therrien, E, Moitessier, N, Roggo, S, Veenstra, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
J.Med.Chem., 48, 2005
|
|
7XNX
 
 | |
7XNY
 
 | |
5GWG
 
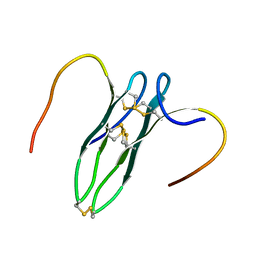 | | Solution structure of rattusin | | Descriptor: | Defensin alpha-related sequence 1 | | Authors: | Lee, C.W, Min, H.J. | | Deposit date: | 2016-09-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rattusin structure reveals a novel defensin scaffold formed by intermolecular disulfide exchanges
Sci Rep, 7, 2017
|
|
5ZTX
 
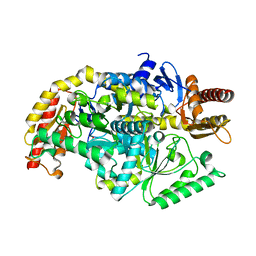 | | co-factor free Transaminase | | Descriptor: | 1,2-ETHANEDIOL, transaminase | | Authors: | Park, H.H, Shin, Y.C. | | Deposit date: | 2018-05-05 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural dynamics of the transaminase active site revealed by the crystal structure of a co-factor free omega-transaminase from Vibrio fluvialis JS17
Sci Rep, 8, 2018
|
|
6IO1
 
 | |
