1MV5
 
 | | Crystal structure of LmrA ATP-binding domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yuan, Y, Chen, H, Patel, D. | | Deposit date: | 2002-09-24 | | Release date: | 2003-12-02 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of LmrA ATP-binding domain reveals the two-site alternating mechanism at molecular level
To be Published
|
|
1OSW
 
 | | The Stem of SL1 RNA in HIV-1: Structure and Nucleocapsid Protein Binding for a 1X3 Internal Loop | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*CP*GP*CP*UP*AP*CP*GP*GP*CP*GP*AP*GP*GP*CP*UP*CP*CP*A)-3' | | Authors: | Yuan, Y, Kerwood, D.J, Paoletti, A.C, Shubsda, M.F, Borer, P.N. | | Deposit date: | 2003-03-20 | | Release date: | 2003-05-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Stem of SL1 RNA in HIV-1: Structure and Nucleocapsid Protein Binding for a 1X3 Internal Loop
Biochemistry, 42, 2003
|
|
6OKY
 
 | | Solution structure of truncated peptide from PAMap53 | | Descriptor: | Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
J.Struct.Biol., 208, 2019
|
|
6OKX
 
 | | Solution structure of VEK50RH1/AA | | Descriptor: | Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
J.Struct.Biol., 208, 2019
|
|
6OQ9
 
 | |
6OQJ
 
 | |
6OKW
 
 | | Solution structure of VEK50 | | Descriptor: | Plasminogen-binding group A streptococcal M-like protein PAM | | Authors: | Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-02-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structural model of the complex of the binding regions of human plasminogen with its M-protein receptor from Streptococcus pyogenes.
J.Struct.Biol., 208, 2019
|
|
6OQK
 
 | |
2OQO
 
 | |
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | Descriptor: | Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
4FMM
 
 | | Dimeric Sec14 family homolog 3 from Saccharomyces cerevisiae presents some novel features of structure that lead to a surprising "dimer-monomer" state change induced by substrate binding | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatidylinositol transfer protein PDR16 | | Authors: | Yuan, Y, Zhao, W, Wang, X, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dimeric Sfh3 has structural changes in its binding pocket that are associated with a dimer-monomer state transformation induced by substrate binding.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3D3H
 
 | | Crystal structure of a complex of the peptidoglycan glycosyltransferase domain from Aquifex aeolicus and neryl moenomycin A | | Descriptor: | (2R)-3-{[(S)-{[(2R,3R,4R,5S,6S)-3-{[(2S,3R,4R,5S,6R)-3-(acetylamino)-5-{[(2S,3R,4R,5S,6R)-3-(acetylamino)-5-{[(2R,3R,4S,5R,6S)-6-carbamoyl-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl]oxy}-4-hydroxy-6-methyltetrahydro-2H-pyran-2-yl]oxy}-4-hydroxy-6-({[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}methyl)tetrahydro-2H-pyran-2-yl]oxy}-6-carbamoyl-4-(carbamoyloxy)-5-hydroxy-5-methyltetrahydro-2H-pyran-2-yl]oxy}(hydroxy)phosphoryl]oxy}-2-{[(2Z)-3,7-dimethylocta-2,6-dien-1-yl]oxy}propanoic acid, Penicillin-insensitive transglycosylase | | Authors: | Yuan, Y, Sliz, P, Walker, S. | | Deposit date: | 2008-05-11 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of the contacts anchoring moenomycin to peptidoglycan glycosyltransferases and implications for antibiotic design.
Acs Chem.Biol., 3, 2008
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
7EW1
 
 | | Cryo-EM structure of siponimod -bound Sphingosine-1-phosphate receptor 5 in complex with Gi protein | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Shao, Z.H, Su, Z.M. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
7EW0
 
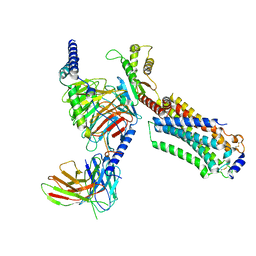 | | Cryo-EM structure of ozanimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | 5-[3-[(1~{S})-1-(2-hydroxyethylamino)-2,3-dihydro-1~{H}-inden-4-yl]-1,2,4-oxadiazol-5-yl]-2-propan-2-yloxy-benzenecarbonitrile, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Su, Z.M, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
7EVZ
 
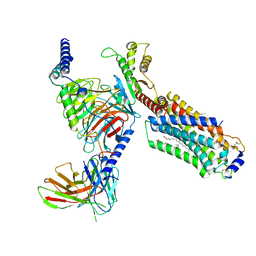 | | Cryo-EM structure of cenerimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | Descriptor: | (2~{S})-3-[4-[5-(2-cyclopentyl-6-methoxy-pyridin-4-yl)-1,2,4-oxadiazol-3-yl]-2-ethyl-6-methyl-phenoxy]propane-1,2-diol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Y, Jia, G.W, Shao, Z.H, Su, Z.M. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
6L8Q
 
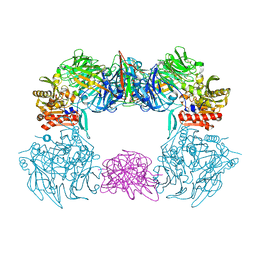 | | Complex structure of bat CD26 and MERS-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yuan, Y. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis of Binding between Middle East Respiratory Syndrome Coronavirus and CD26 from Seven Bat Species.
J.Virol., 94, 2020
|
|
5V4U
 
 | |
5Z2H
 
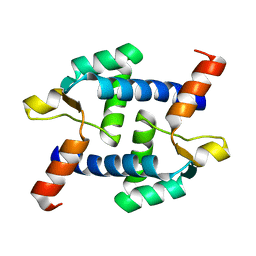 | | Structure of Dictyostelium discoideum mitochondrial calcium uniporter N-terminal domain(DdMCU-NTD) | | Descriptor: | Dictyostelium discoideum mitochondrial calcium uniporter | | Authors: | Yuan, Y, Wen, M, Chou, J.J, Li, D, Bo, O. | | Deposit date: | 2018-01-02 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.674 Å) | | Cite: | Structural Characterization of the N-Terminal Domain of theDictyostelium discoideumMitochondrial Calcium Uniporter.
Acs Omega, 5, 2020
|
|
5Z2I
 
 | | Structure of Dictyostelium discoideum mitochondrial calcium uniporter N-ternimal domain (Se-DdMCU-NTD) | | Descriptor: | Dictyostelium discoideum mitochondrial calcium uniporter | | Authors: | Yuan, Y, Wen, M, Chou, J, Li, D, Bo, O. | | Deposit date: | 2018-01-02 | | Release date: | 2019-01-02 | | Last modified: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural Characterization of the N-Terminal Domain of theDictyostelium discoideumMitochondrial Calcium Uniporter.
Acs Omega, 5, 2020
|
|
