1KHI
 
 | | CRYSTAL STRUCTURE OF HEX1 | | Descriptor: | Hex1 | | Authors: | Yuan, P, Swaminathan, K. | | Deposit date: | 2001-11-30 | | Release date: | 2002-11-30 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa
NAT.STRUCT.BIOL., 10, 2003
|
|
3BVP
 
 | |
3EBJ
 
 | | Crystal structure of an avian influenza virus protein | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein | | Authors: | Yuan, P, Bartlam, M, Lou, Z, Chen, S, Rao, Z, Liu, Y. | | Deposit date: | 2008-08-27 | | Release date: | 2009-02-10 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site
Nature, 458, 2009
|
|
1Z4X
 
 | | Parainfluenza Virus 5 (SV5) Hemagglutinin-Neuraminidase (HN) with ligand Sialyllactose (soaked with Sialyllactose, pH8.0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yuan, P, Thompson, T.B, Wurzburg, B.A, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose.
Structure, 13, 2005
|
|
1Z4V
 
 | | Parainfluenza Virus 5 (SV5) Hemagglutinin-Neuraminidase (HN) with ligand DANA (soaked with DANA, pH 7.0) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, P, Thompson, T.B, Wurzburg, B.A, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose.
Structure, 13, 2005
|
|
1Z4Z
 
 | | Parainfluenza Virus 5 (SV5) Hemagglutinin-Neuraminidase (HN) with ligand DANA(soaked with sialic acid, pH7.0)) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, P, Thompson, T.B, Wurzburg, B.A, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose.
Structure, 13, 2005
|
|
1Z4W
 
 | | Parainfluenza Virus 5 (SV5) Hemagglutinin-Neuraminidase (HN) with ligand DANA (soaked with DANA, pH8.0) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, P, Thompson, T.B, Wurzburg, B.A, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose.
Structure, 13, 2005
|
|
1Z50
 
 | | Parainfluenza Virus 5 (SV5) Hemagglutinin-Neuraminidase (HN) with ligand DANA (soaked with sialic acid, pH 8.0) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, P, Thompson, T.B, Wurzburg, B.A, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose.
Structure, 13, 2005
|
|
1Z4Y
 
 | | Parainfluenza Virus 5 (SV5) Hemagglutinin-Neuraminidase (HN) (pH 8.0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yuan, P, Thompson, T.B, Wurzburg, B.A, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose.
Structure, 13, 2005
|
|
3MT5
 
 | | Crystal Structure of the Human BK Gating Apparatus | | Descriptor: | CALCIUM ION, Potassium large conductance calcium-activated channel, subfamily M, ... | | Authors: | Yuan, P, MacKinnon, R. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the human BK channel Ca2+-activation apparatus at 3.0 A resolution.
Science, 329, 2010
|
|
3T1E
 
 | | The structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a 4-helix bundle stalk | | Descriptor: | Hemagglutinin-neuraminidase | | Authors: | Yuan, P, Swanson, K, Leser, G.P, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2011-07-21 | | Release date: | 2011-09-07 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U6N
 
 | | Open Structure of the BK channel Gating Ring | | Descriptor: | CALCIUM ION, High-Conductance Ca2+-Activated K+ Channel protein | | Authors: | Yuan, P, MacKinnon, R. | | Deposit date: | 2011-10-12 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Open structure of the Ca(2+) gating ring in the high-conductance Ca(2+)-activated K(+) channel.
Nature, 481, 2011
|
|
4FZH
 
 | | Structure of the Ulster Strain Newcastle Disease Virus Hemagglutinin-Neuraminidase Reveals Auto-Inhibitory Interactions Associated with Low Virulence | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-neuraminidase | | Authors: | Yuan, P, Paterson, R.G, Leser, G.P, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2012-07-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5008 Å) | | Cite: | Structure of the ulster strain newcastle disease virus hemagglutinin-neuraminidase reveals auto-inhibitory interactions associated with low virulence.
Plos Pathog., 8, 2012
|
|
1UII
 
 | |
7SI7
 
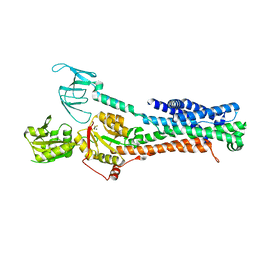 | | Structure of ATP7B in state 2 | | Descriptor: | MAGNESIUM ION, P-type Cu(+) transporter, TETRAFLUOROALUMINATE ION | | Authors: | Bitter, R.M, Oh, S.C, Hite, R.K, Yuan, P. | | Deposit date: | 2021-10-12 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the Wilson disease copper transporter ATP7B.
Sci Adv, 8, 2022
|
|
7SI3
 
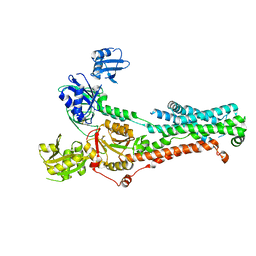 | | Consensus structure of ATP7B | | Descriptor: | MAGNESIUM ION, P-type Cu(+) transporter, TETRAFLUOROALUMINATE ION | | Authors: | Bitter, R.M, Oh, S.C, Hite, R.K, Yuan, P. | | Deposit date: | 2021-10-12 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure of the Wilson disease copper transporter ATP7B.
Sci Adv, 8, 2022
|
|
7SI6
 
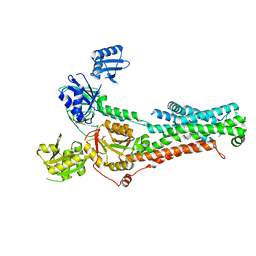 | | Structure of ATP7B in state 1 | | Descriptor: | MAGNESIUM ION, P-type Cu(+) transporter, TETRAFLUOROALUMINATE ION | | Authors: | Bitter, R.M, Oh, S.C, Hite, R.K, Yuan, P. | | Deposit date: | 2021-10-12 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structure of the Wilson disease copper transporter ATP7B.
Sci Adv, 8, 2022
|
|
8F7C
 
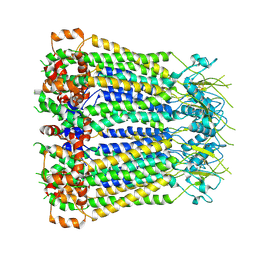 | | Cryo-EM structure of human pannexin 2 | | Descriptor: | Pannexin-2, Soluble cytochrome b562 fusion | | Authors: | He, Z, Yuan, P. | | Deposit date: | 2022-11-18 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural and functional analysis of human pannexin 2 channel.
Nat Commun, 14, 2023
|
|
6UW6
 
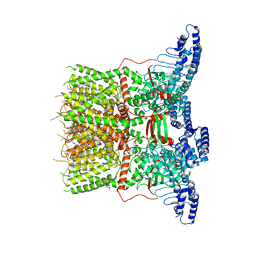 | | Cryo-EM structure of the human TRPV3 K169A mutant determined in lipid nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 3, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Gating of human TRPV3 in a lipid bilayer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UW9
 
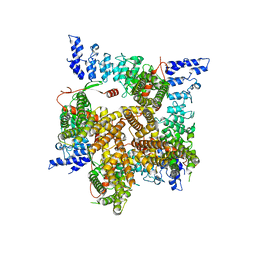 | |
6UW4
 
 | | Cryo-EM structure of human TRPV3 determined in lipid nanodisc | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily V member 3, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Gating of human TRPV3 in a lipid bilayer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6V6D
 
 | | Cryo-EM structure of human pannexin 1 | | Descriptor: | Pannexin-1 | | Authors: | Deng, Z, He, Z, Yuan, P. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-01 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Cryo-EM structures of the ATP release channel pannexin 1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UW8
 
 | |
6UZY
 
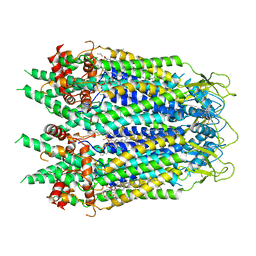 | | Cryo-EM structure of Xenopus tropicalis pannexin 1 | | Descriptor: | HEXADECANE, Pannexin, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, He, Z, Yuan, P. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-01 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of the ATP release channel pannexin 1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6V5A
 
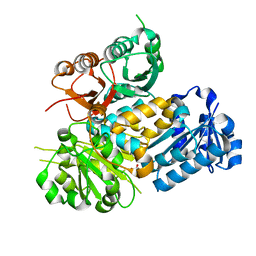 | | Crystal structure of the human BK channel gating ring L390P mutant | | Descriptor: | CALCIUM ION, Calcium-activated potassium channel subunit alpha-1, SULFATE ION | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-12-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coupling of Ca2+and voltage activation in BK channels through the alpha B helix/voltage sensor interface.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
