2VHE
 
 | | PglD-CoA complex: An acetyl transferase from Campylobacter jejuni | | Descriptor: | ACETYLTRANSFERASE, COENZYME A, SULFATE ION | | Authors: | Rangarajan, E.S, Ruane, K.M, Sulea, T, Watson, D.C, Proteau, A, Leclerc, S, Cygler, M, Matte, A, Young, N.M. | | Deposit date: | 2007-11-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Active Site Residues of Pgld, an N-Acetyltransferase from the Bacillosamine Synthetic Pathway Required for N-Glycan Synthesis in Campylobacter Jejuni
Biochemistry, 47, 2008
|
|
1JV5
 
 | | Anti-blood group A Fv | | Descriptor: | Ig chain heavy chain precursor V region, Ig kappa chain precursor V region | | Authors: | Thomas, R, Patenaude, S.I, MacKenzie, C.R, To, R, Hirama, T, Young, N.M, Evans, S.V. | | Deposit date: | 2001-08-28 | | Release date: | 2002-01-09 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an anti-blood group A Fv and improvement of its binding affinity without loss of specificity.
J.Biol.Chem., 277, 2002
|
|
3BFP
 
 | | Crystal Structure of apo-PglD from Campylobacter jejuni | | Descriptor: | Acetyltransferase, CITRATE ANION | | Authors: | Rangarajan, E.S, Watson, D.C, Leclerc, S, Proteau, A, Cygler, M, Matte, A, Young, N.M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-11-22 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Active Site Residues of PglD, an N-Acetyltransferase from the Bacillosamine Synthetic Pathway Required for N-Glycan Synthesis in Campylobacter jejuni.
Biochemistry, 47, 2008
|
|
3LM1
 
 | | Crystal Structure Analysis of Maclura pomifera agglutinin complex with p-nitrophenyl-GalNAc | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Agglutinin alpha chain, Agglutinin beta-2 chain | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
1JOT
 
 | | STRUCTURE OF THE LECTIN MPA COMPLEXED WITH T-ANTIGEN DISACCHARIDE | | Descriptor: | AGGLUTININ, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Lee, X, Thompson, A, Zhang, Z, Hoa, T.-T, Biesterfeldt, J, Ogata, C, Xu, L, Johnston, R.A.Z, Young, N.M. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of Maclura pomifera agglutinin and the T-antigen disaccharide, Galbeta1,3GalNAc.
J.Biol.Chem., 273, 1998
|
|
1JZN
 
 | | crystal structure of a galactose-specific C-type lectin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Galactose-specific lectin, ... | | Authors: | Walker, J.R, Nagar, B, Young, N.M, Hirama, T, Rini, J.M. | | Deposit date: | 2001-09-16 | | Release date: | 2003-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Crystal Structure of a Galactose-Specific C-Type Lectin Possessing a Novel Decameric Quaternary Structure.
Biochemistry, 43, 2004
|
|
4ZTC
 
 | | PglE Aminotransferase in complex with External Aldimine, Mutant K184A | | Descriptor: | Aminotransferase homolog, [(2R,3R,4R,5S,6R)-3-acetamido-6-methyl-5-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-4-oxidanyl-oxan-2-yl] [[(2R,3S,4R,5R)-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] hydrogen phosphate | | Authors: | Riegert, A.S, Thoden, J.B, Young, N.M, Watson, D.C, Holden, H.M. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the external aldimine form of PglE, an aminotransferase required for N,N'-diacetylbacillosamine biosynthesis.
Protein Sci., 24, 2015
|
|
3LLY
 
 | | Crystal Structure Analysis of Maclura pomifera agglutinin | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-2 chain | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
3LLZ
 
 | | Crystal Structure Analysis of Maclura pomifera agglutinin complex with Gal-beta-1,3-GalNAc | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-2 chain, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
1MUQ
 
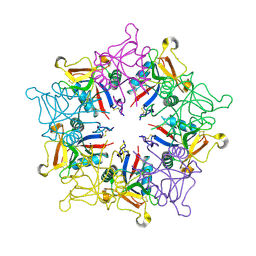 | | X-ray Crystal Structure of Rattlesnake Venom Complexed With Thiodigalactoside | | Descriptor: | 1-thio-beta-D-galactopyranose-(1-1)-beta-D-galactopyranose, CALCIUM ION, Galactose-specific lectin, ... | | Authors: | Walker, J.R, Nagar, B, Young, N.M, Hirama, T, Rini, J.M. | | Deposit date: | 2002-09-24 | | Release date: | 2003-07-01 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure of a Galactose-Specific C-Type Lectin Possessing a Novel Decameric Quaternary Structure.
Biochemistry, 43, 2004
|
|
1PLG
 
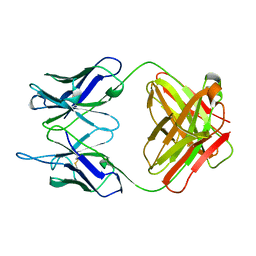 | | EVIDENCE FOR THE EXTENDED HELICAL NATURE OF POLYSACCHARIDE EPITOPES. THE 2.8 ANGSTROMS RESOLUTION STRUCTURE AND THERMODYNAMICS OF LIGAND BINDING OF AN ANTIGEN BINDING FRAGMENT SPECIFIC FOR ALPHA-(2->8)-POLYSIALIC ACID | | Descriptor: | IGG2A=KAPPA= | | Authors: | Evans, S.V, Sigurskjold, B.W, Jennings, H.J, Brisson, J.-R, Tse, W.C, To, R, Altman, E, Frosch, M, Weisgerber, C, Kratzin, H, Klebert, S, Vaesen, M, Bitter-Suermann, D, Rose, D.R, Young, N.M, Bundle, D.R. | | Deposit date: | 1995-04-24 | | Release date: | 1996-04-03 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the extended helical nature of polysaccharide epitopes. The 2.8 A resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for alpha-(2-->8)-polysialic acid.
Biochemistry, 34, 1995
|
|
2HXW
 
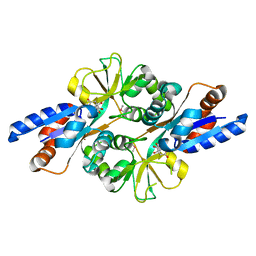 | | Crystal Structure of Peb3 from Campylobacter jejuni | | Descriptor: | CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Rangarajan, E.S, Bhatia, S, Watson, D.C, Munger, C, Cygler, M, Matte, A, Young, N.M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-08-04 | | Release date: | 2007-05-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural context for protein N-glycosylation in bacteria: The structure of PEB3, an adhesin from Campylobacter jejuni.
Protein Sci., 16, 2007
|
|
1MFA
 
 | |
5KXD
 
 | | Wisteria floribunda lectin in complex with GalNAc(beta1-4)GlcNAc (LacdiNAc) at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Evans, S.V, Haji-Ghassemi, O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis for Recognition of the Cancer Glycobiomarker, LacdiNAc (GalNAc[ beta 14]GlcNAc), by Wisteria floribunda Agglutinin.
J.Biol.Chem., 291, 2016
|
|
5KXC
 
 | | Wisteria floribunda lectin in complex with GalNAc(beta1-4)GlcNAc (LacdiNAc) at pH 8.5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Evans, S.V, Haji-Ghassemi, O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Recognition of the Cancer Glycobiomarker, LacdiNAc (GalNAc[ beta 14]GlcNAc), by Wisteria floribunda Agglutinin.
J.Biol.Chem., 291, 2016
|
|
5KXB
 
 | | Wisteria floribunda lectin in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Evans, S.V, Haji-Ghassemi, O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular Basis for Recognition of the Cancer Glycobiomarker, LacdiNAc (GalNAc[ beta 14]GlcNAc), by Wisteria floribunda Agglutinin.
J.Biol.Chem., 291, 2016
|
|
5KXE
 
 | | Wisteria floribunda lectin in complex with GalNAc(beta1-4)GlcNAc (LacdiNAc) at pH 4.2 | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Evans, S.V, Haji-Ghassemi, O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Molecular Basis for Recognition of the Cancer Glycobiomarker, LacdiNAc (GalNAc[ beta 14]GlcNAc), by Wisteria floribunda Agglutinin.
J.Biol.Chem., 291, 2016
|
|
1MAM
 
 | |
3FJ7
 
 | | Crystal structure of L-phospholactate Bound PEB3 | | Descriptor: | L-PHOSPHOLACTATE, Major antigenic peptide PEB3 | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
3FJG
 
 | | Crystal structure of 3PG bound PEB3 | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Major antigenic peptide PEB3 | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
3FJM
 
 | | crystal structure of phosphate bound PEB3 | | Descriptor: | Major antigenic peptide PEB3, PHOSPHATE ION | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
3FIR
 
 | | Crystal structure of Glycosylated K135E PEB3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-L-glucopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Min, T, Matte, A, Cygler, M. | | Deposit date: | 2008-12-12 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes.
Biochemistry, 48, 2009
|
|
5BJV
 
 | | X-ray structure of the PglF UDP-N-acetylglucosamine 4,6-dehydratase from Campylobacterjejuni, D396N/K397A variant in complex with UDP-N-acrtylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BJU
 
 | | X-ray structure of the PglF dehydratase from Campylobacter jejuni in complex with UDP and NAD(H) | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
5BJW
 
 | | X-ray structure of the PglF 4,6-dehydratase from campylobacter jejuni, T595S variant, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
