7FGP
 
 | |
7Y9P
 
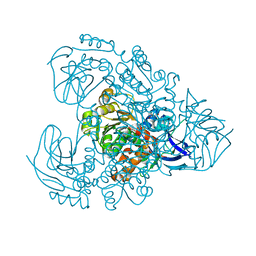 | | Xylitol dehydrogenase S96C/S99C/Y102C mutant(thermostabilized form) from Pichia stipitis | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Yoshiwara, K, Watanabe, Y, Watanabe, S. | | Deposit date: | 2022-06-25 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular evolutionary insight of structural zinc atom in yeast xylitol dehydrogenases and its application in bioethanol production by lignocellulosic biomass.
Sci Rep, 13, 2023
|
|
7B81
 
 | |
7CGR
 
 | |
7CGQ
 
 | |
7DO6
 
 | |
7DO5
 
 | |
7DO7
 
 | |
7WWX
 
 | | Crystal structure of Herbaspirillum huttiense L-arabinose 1-dehydrogenase (NAD bound form) | | Descriptor: | DI(HYDROXYETHYL)ETHER, NAD(P)-dependent dehydrogenase (Short-subunit alcohol dehydrogenase family), NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Matsubara, R, Yoshiwara, K, Watanabe, Y, Watanabe, S. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of L-arabinose 1-dehydrogenase as a short-chain reductase/dehydrogenase protein.
Biochem.Biophys.Res.Commun., 604, 2022
|
|
