7DBT
 
 | | Crystal structure of catalytic domain of Anhydrobiosis-related Mn-dependent Peroxidase (AMNP) from Ramazzottius varieornatus (Mn2+-bound form) | | Descriptor: | AMNP/g12777, MANGANESE (II) ION | | Authors: | Yoshida, Y, Satoh, T, Ota, C, Tanaka, S, Horikawa, D.D, Tomita, M, Kato, K, Arakawa, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-series transcriptomic screening of factors contributing to the cross-tolerance to UV radiation and anhydrobiosis in tardigrades.
Bmc Genomics, 23, 2022
|
|
7DBU
 
 | | Crystal structure of catalytic domain of Anhydrobiosis-related Mn-dependent Peroxidase (AMNP) from Ramazzottius varieornatus (Zn2+-bound form) | | Descriptor: | AMNP/g12777, ZINC ION | | Authors: | Yoshida, Y, Satoh, T, Ota, C, Tanaka, S, Horikawa, D.D, Tomita, M, Kato, K, Arakawa, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-10-06 | | Last modified: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-series transcriptomic screening of factors contributing to the cross-tolerance to UV radiation and anhydrobiosis in tardigrades.
Bmc Genomics, 23, 2022
|
|
6IOY
 
 | | Crystal structure of Porphyromonas gingivalis acetate kinase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Acetate kinase, SULFATE ION | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2018-10-31 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of the phosphotransacetylase-acetate kinase pathway for ATP production inPorphyromonas gingivalis.
J Oral Microbiol, 11, 2019
|
|
6IOX
 
 | |
6IOW
 
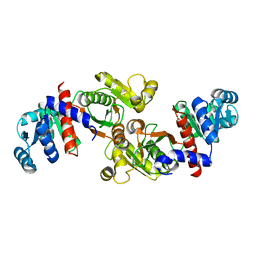 | |
5XEM
 
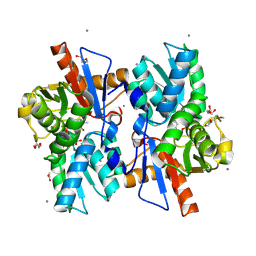 | |
3B1E
 
 | | Crystal structure of betaC-S lyase from Streptococcus anginosus in complex with L-serine: alpha-Aminoacrylate form | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into catalysis by beta C-S lyase from Streptococcus anginosus
Proteins, 80, 2012
|
|
3B1C
 
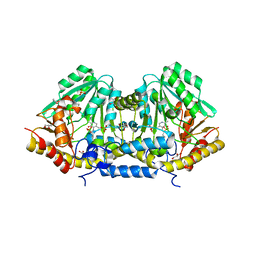 | | Crystal structure of betaC-S lyase from Streptococcus anginosus: Internal aldimine form | | Descriptor: | BetaC-S lyase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-27 | | Last modified: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into catalysis by beta C-S lyase from Streptococcus anginosus
Proteins, 80, 2012
|
|
3B1D
 
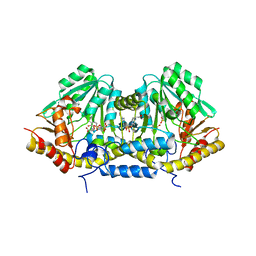 | | Crystal structure of betaC-S lyase from Streptococcus anginosus in complex with L-serine: External aldimine form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BetaC-S lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural insights into catalysis by beta C-S lyase from Streptococcus anginosus
Proteins, 80, 2012
|
|
2RJ2
 
 | | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution | | Descriptor: | CHLORIDE ION, F-box only protein 2, NICKEL (II) ION | | Authors: | Vaijayanthimala, S, Velmurugan, D, Mizushima, T, Yamane, T, Yoshida, Y, Tanaka, K. | | Deposit date: | 2007-10-14 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Sugar Recognizing SCF Ubiquitin Ligase at 1.7 Resolution
To be Published
|
|
2NLI
 
 | | Crystal Structure of the complex between L-lactate oxidase and a substrate analogue at 1.59 angstrom resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, HYDROGEN PEROXIDE, LACTIC ACID, ... | | Authors: | Furuichi, M, Suzuki, N, Balasundaresan, D, Yoshida, Y, Minagawa, H, Watanabe, Y, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2006-10-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
2ZFA
 
 | | Structure of Lactate Oxidase at pH4.5 from AEROCOCCUS VIRIDANS | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Lactate oxidase | | Authors: | Furuichi, M, Balasundaresan, D, Suzuki, N, Yoshida, Y, Minagawa, H, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
5B53
 
 | |
5B55
 
 | | Crystal structure of hydrogen sulfide-producing enzyme (Fn1055) D232N mutant in complexed with alpha-aminoacrylate intermediate: lysine-dimethylated form | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Cysteine synthase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2016-04-22 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural insights into the catalytic mechanism of cysteine (hydroxyl) lyase from the hydrogen sulfide-producing oral pathogen,Fusobacterium nucleatum.
Biochem. J., 475, 2018
|
|
5XEO
 
 | |
5XEN
 
 | |
1J1G
 
 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1J1F
 
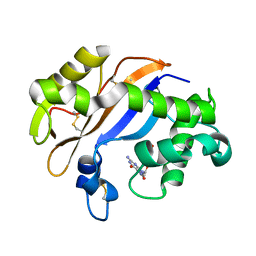 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
5B4N
 
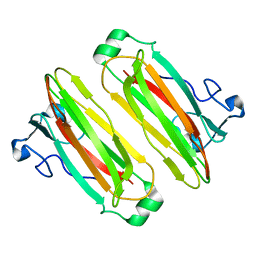 | | Structure analysis of function associated loop mutant of substrate recognition domain of Fbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2 | | Authors: | Nishio, K, Yoshida, Y, Tanaka, K, Mizushima, T. | | Deposit date: | 2016-04-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a function-associated loop mutant of the substrate-recognition domain of Fbs1 ubiquitin ligase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5Z5C
 
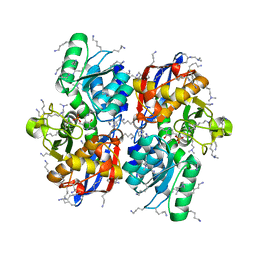 | | Crystal structure of hydrogen sulfide-producing enzyme (Fn1055) from Fusobacterium nucleatum: lysine-dimethylated form | | Descriptor: | CHLORIDE ION, Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kezuka, Y, Yoshida, Y, Nonaka, T. | | Deposit date: | 2018-01-17 | | Release date: | 2018-02-14 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into the catalytic mechanism of cysteine (hydroxyl) lyase from the hydrogen sulfide-producing oral pathogen,
Biochem. J., 475, 2018
|
|
1UMI
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, F-box only protein 2 | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K. | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UMH
 
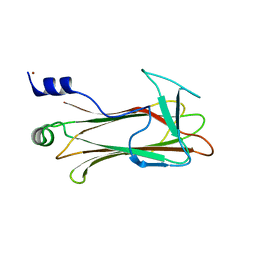 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | F-box only protein 2, NICKEL (II) ION | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
2D35
 
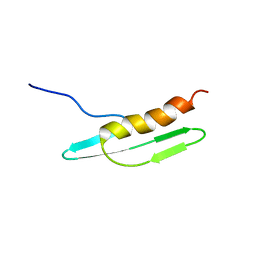 | | Solution structure of Cell Division Reactivation Factor, CedA | | Descriptor: | Cell division activator cedA | | Authors: | Abe, Y, Watanabe, N, Matsuda, Y, Yoshida, Y, Katayama, T, Ueda, T. | | Deposit date: | 2005-09-26 | | Release date: | 2006-12-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis and Molecular Interaction of Cell Division Reactivation Factor, CedA from Escherichia coli
To be Published
|
|
2E33
 
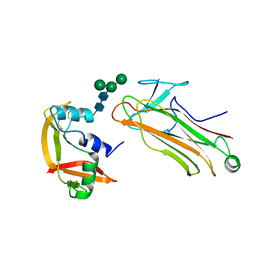 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, Ribonuclease pancreatic, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E31
 
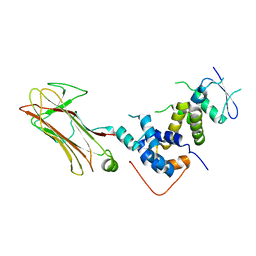 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, S-phase kinase-associated protein 1A | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
