4Q0Q
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribulose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0U
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0S
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with ribitol | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), D-ribitol, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0P
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0V
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribulose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
8GRJ
 
 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone
To Be Published
|
|
4GJI
 
 | |
4GJJ
 
 | |
3M0X
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329L in complex with D-psicose | | Descriptor: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0Y
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329A in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0H
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0L
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with D-psicose | | Descriptor: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0V
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329L in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M0M
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with D-allose | | Descriptor: | D-ALLOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3ITY
 
 | | Metal-free form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITO
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant D327N in complex with D-psicose | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION, alpha-D-psicofuranose | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITT
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329K in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IUD
 
 | | Cu2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | COPPER (II) ION, L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITL
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant D327N in complex with L-rhamnulose | | Descriptor: | 6-deoxy-beta-L-fructofuranose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IUI
 
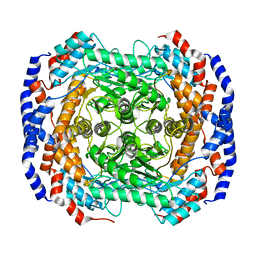 | | Zn2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITX
 
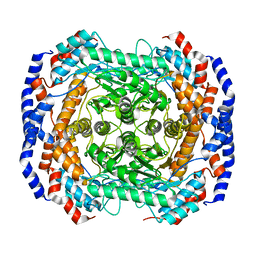 | | Mn2+ bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITV
 
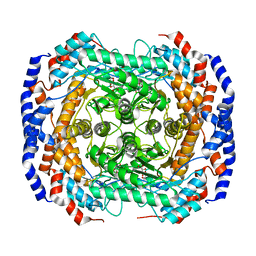 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329K in complex with D-psicose | | Descriptor: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IUH
 
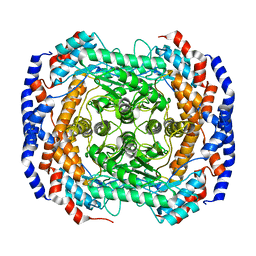 | | Co2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | COBALT (II) ION, L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
5J8L
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy L-tagatose, using a crystal grown in microgravity | | Descriptor: | 1-deoxy-L-tagatose, 1-deoxy-beta-L-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Izumori, K, Kamitori, S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
8JQ3
 
 | |
