5HPP
 
 | |
1AUM
 
 | | HIV CAPSID C-TERMINAL DOMAIN (CAC146) | | Descriptor: | HIV CAPSID | | Authors: | Hill, C.P, Gamble, T.R, Yoo, S, Vajdos, F.F, Von Schwedler, U.K, Worthylake, D.K, Wang, H, Mccutcheon, J.P, Sundquist, W.I. | | Deposit date: | 1997-08-29 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein.
Science, 278, 1997
|
|
1A8O
 
 | | HIV CAPSID C-TERMINAL DOMAIN | | Descriptor: | HIV CAPSID | | Authors: | Gamble, T.R, Yoo, S, Vajdos, F.F, Von Schwedler, U.K, Worthylake, D.K, Wang, H, Mccutcheon, J.P, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein.
Science, 278, 1997
|
|
1A43
 
 | | STRUCTURE OF THE HIV-1 CAPSID PROTEIN DIMERIZATION DOMAIN AT 2.6A RESOLUTION | | Descriptor: | HIV-1 CAPSID | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-02-10 | | Release date: | 1999-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BAJ
 
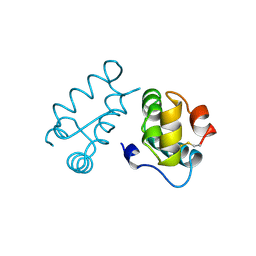 | | HIV-1 CAPSID PROTEIN C-TERMINAL FRAGMENT PLUS GAG P2 DOMAIN | | Descriptor: | GAG POLYPROTEIN | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5SUS
 
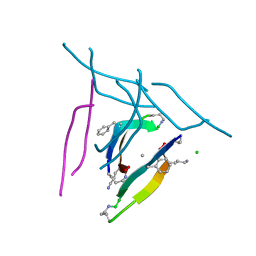 | | X-ray crystallographic structure of a covalent trimer derived from A-beta 17_36. X-ray diffractometer data set. (ORN)CVF(MEA)CED(ORN)AIIGL(ORN)V. | | Descriptor: | 16mer A-beta peptide: ORN-CYS-VAL-PHE-MEA-CYS-GLU-ASP-ORN-ALA-ILE-ILE-GLY-LEU-ORN-VAL, CHLORIDE ION, SODIUM ION | | Authors: | Kreutzer, A.G, Yoo, S, Nowick, J.S. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Stabilization, Assembly, and Toxicity of Trimers Derived from A beta.
J.Am.Chem.Soc., 139, 2017
|
|
5SUR
 
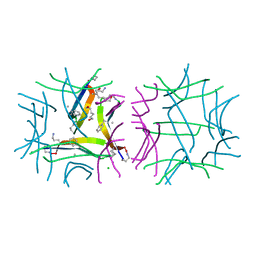 | | X-ray crystallographic structure of a covalent trimer derived from A-beta 17_36. Synchrotron data set. (ORN)CVF(MEA)CED(ORN)AIIGL(ORN)V. | | Descriptor: | 16mer A-beta peptide: ORN-CYS-VAL-PHE-MEA-CYS-GLU-ASP-ORN-ALA-ILE-ILE-GLY-LEU-ORN-VAL, CHLORIDE ION, HEXANE-1,6-DIOL, ... | | Authors: | Kreutzer, A.G, Yoo, S, Nowick, J.S. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Stabilization, Assembly, and Toxicity of Trimers Derived from A beta.
J.Am.Chem.Soc., 139, 2017
|
|
8EC9
 
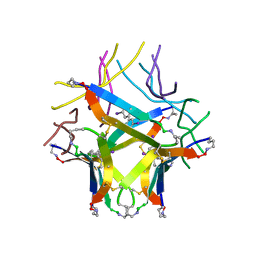 | |
8ECA
 
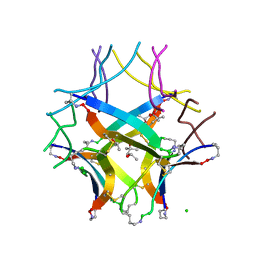 | | Covalently stabilized triangular trimer derived from Abeta16-36 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ORN-LYS-LEU-VAL-PHE-PHE-ALA-GLU-ORN-CYS-ILE-ILE-SAR-CYS-MET | | Authors: | Kreutzer, A.G, Yoo, S, Nowick, J.S. | | Deposit date: | 2022-09-01 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Probing differences among A beta oligomers with two triangular trimers derived from A beta.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1GWP
 
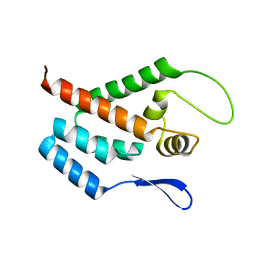 | | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE MATURE HIV-1 CAPSID PROTEIN | | Descriptor: | GAG POLYPROTEIN | | Authors: | Tang, C, Gitti, R.K, Lee, B.M, Walker, J, Summers, M.F, Yoo, S, Sundquist, W.I. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-21 | | Last modified: | 2014-03-12 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal 283-Residue Fragment of the Immature HIV-1 Gag Polyprotein
Nat.Struct.Biol., 9, 2002
|
|
1AWU
 
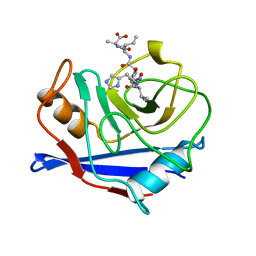 | | CYPA COMPLEXED WITH HVGPIA (PSEUDO-SYMMETRIC MONOMER) | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-05 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1AWV
 
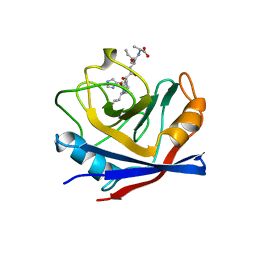 | | CYPA COMPLEXED WITH HVGPIA | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-05 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1AK4
 
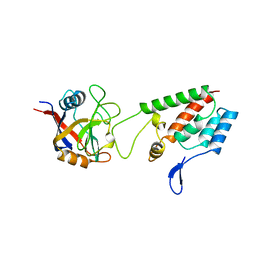 | | HUMAN CYCLOPHILIN A BOUND TO THE AMINO-TERMINAL DOMAIN OF HIV-1 CAPSID | | Descriptor: | CYCLOPHILIN A, HIV-1 CAPSID | | Authors: | Hill, C.P, Gamble, T.R, Vajdos, F.F, Worthylake, D.K, Sundquist, W.I. | | Deposit date: | 1997-05-28 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid.
Cell(Cambridge,Mass.), 87, 1996
|
|
1AWT
 
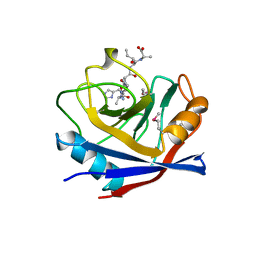 | | SECYPA COMPLEXED WITH HAGPIA | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-05 | | Release date: | 1998-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1AWR
 
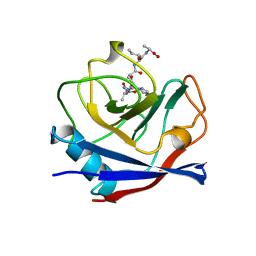 | | CYPA COMPLEXED WITH HAGPIA | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-04 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1AWS
 
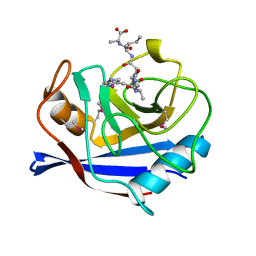 | | SECYPA COMPLEXED WITH HAGPIA (PSEUDO-SYMMETRIC MONOMER) | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-04 | | Release date: | 1998-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1AWQ
 
 | | CYPA COMPLEXED WITH HAGPIA (PSEUDO-SYMMETRIC MONOMER) | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-04 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
6DR5
 
 | |
6DR4
 
 | |
6DR6
 
 | |
4F3L
 
 | | Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex | | Descriptor: | BMAL1b, Circadian locomoter output cycles protein kaput | | Authors: | Huang, N, Chelliah, Y, Shan, Y, Taylor, C, Yoo, S, Partch, C, Green, C.B, Zhang, H, Takahashi, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex.
Science, 337, 2012
|
|
5SUT
 
 | | X-ray crystallographic structure of a covalent trimer derived from A-beta 17_36. Synchrotron data set. (ORN)CVFFCED(ORN)AII(SAR)L(ORN)V. | | Descriptor: | 16mer A-beta peptide: ORN-CYS-VAL-PHE-PHE-CYS-GLU-ASP-ORN-ALA-ILE-ILE-SAR-LEU-ORN-VAL, CHLORIDE ION | | Authors: | Kreutzer, A.G, Spencer, R.K, Nowick, J.S. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Stabilization, Assembly, and Toxicity of Trimers Derived from A beta.
J.Am.Chem.Soc., 139, 2017
|
|
5SUU
 
 | | X-ray crystallographic structure of a covalent trimer derived from A-beta 17-36. X-ray diffractometer data set. (ORN)CVFFCED(ORN)AII(SAR)L(ORN)V. | | Descriptor: | 16mer A-beta peptide: ORN-CYS-VAL-PHE-PHE-CYS-GLU-ASP-ORN-ALA-ILE-ILE-SAR-LEU-ORN-VAL, CHLORIDE ION, IODIDE ION | | Authors: | Kreutzer, A.G, Spencer, R.K, Nowick, J.S. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Stabilization, Assembly, and Toxicity of Trimers Derived from A beta.
J.Am.Chem.Soc., 139, 2017
|
|
5W4H
 
 | |
5W4I
 
 | |
