4GB3
 
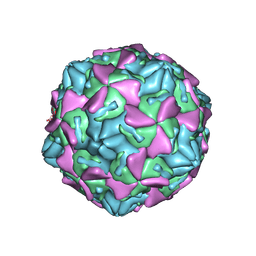 | | Human coxsackievirus B3 strain RD coat protein | | Descriptor: | MYRISTIC ACID, PALMITIC ACID, coat protein 1, ... | | Authors: | Yoder, J.D, Hafenstein, S. | | Deposit date: | 2012-07-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The Crystal Structure of a Coxsackievirus B3-RD Variant and a Refined 9-Angstrom Cryo-Electron Microscopy Reconstruction of the Virus Complexed with Decay-Accelerating Factor (DAF) Provide a New Footprint of DAF on the Virus Surface.
J.Virol., 86, 2012
|
|
2B4I
 
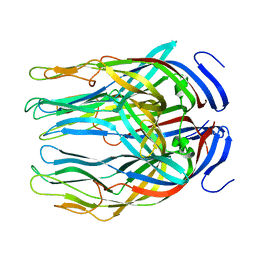 | |
2B4H
 
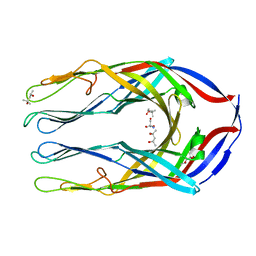 | |
3J24
 
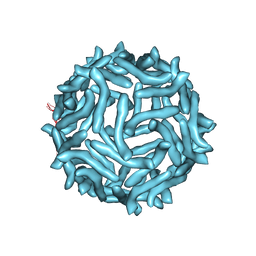 | | CryoEM reconstruction of complement decay-accelerating factor | | Descriptor: | Complement decay-accelerating factor | | Authors: | Yoder, J.D, Hafenstein, S.H. | | Deposit date: | 2012-08-17 | | Release date: | 2012-09-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | The Crystal Structure of a Coxsackievirus B3-RD Variant and a Refined 9-Angstrom Cryo-Electron Microscopy Reconstruction of the Virus Complexed with Decay-Accelerating Factor (DAF) Provide a New Footprint of DAF on the Virus Surface.
J.Virol., 86, 2012
|
|
4GMP
 
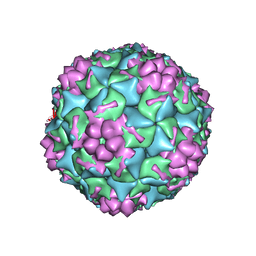 | |
3J7E
 
 | | Electron cryo-microscopy of human papillomavirus 16 and H16.V5 Fab fragments | | Descriptor: | H16.V5 Fab heavy chain, H16.V5 Fab light chain | | Authors: | Lee, H, Brendle, S.A, Bywaters, S.M, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2014-06-23 | | Release date: | 2014-11-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | A cryo-electron microscopy study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment.
J.Virol., 89, 2015
|
|
3J7G
 
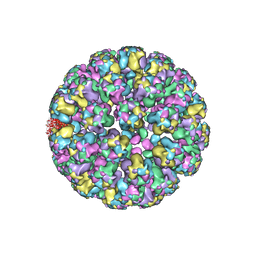 | |
