2BN6
 
 | | P-Element Somatic Inhibitor Protein | | Descriptor: | PSI | | Authors: | Ignjatovic, T, Yang, J.C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
2IGG
 
 | | DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR | | Descriptor: | PROTEIN G | | Authors: | Lian, L.-Y, Derrick, J.P, Sutcliffe, M.J, Yang, J.C, Roberts, G.C.K. | | Deposit date: | 1992-08-26 | | Release date: | 1994-01-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
J.Mol.Biol., 228, 1992
|
|
2IGH
 
 | | DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR | | Descriptor: | PROTEIN G | | Authors: | Lian, L.-Y, Derrick, J.P, Sutcliffe, M.J, Yang, J.C, Roberts, G.C.K. | | Deposit date: | 1992-08-26 | | Release date: | 1994-01-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
J.Mol.Biol., 228, 1992
|
|
7AAB
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with inhibitor EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AAA
 
 | | Crystal structure of the catalytic domain of human PARP1 (apo) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Underwood, E, Rawlins, P.B, Johannes, J.W, Easton, L.E, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AAD
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AAC
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with veliparib | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
5NR6
 
 | |
5NR5
 
 | |
6RH6
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for the complex of NECAP1 PHear domain with phosphorylated AP2 mu2 148-163 | | Descriptor: | AP-2 complex subunit mu, Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6RH5
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for NECAP1 PHear domain | | Descriptor: | Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6TL0
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for the complex of VPS29 with VARP 687-747 | | Descriptor: | Ankyrin repeat domain-containing protein 27, Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Crawley-Snowdon, H. | | Deposit date: | 2019-11-29 | | Release date: | 2020-10-21 | | Method: | SOLUTION NMR | | Cite: | Mechanism and evolution of the Zn-fingernail required for interaction of VARP with VPS29.
Nat Commun, 11, 2020
|
|
1HF9
 
 | | C-Terminal Coiled-Coil Domain from Bovine IF1 | | Descriptor: | ATPASE INHIBITOR (MITOCHONDRIAL) | | Authors: | Gordon-Smith, D.J, Carbajo, R.J, Yang, J.-C, Videler, H, Runswick, M.J, Walker, J.E, Neuhaus, D. | | Deposit date: | 2000-11-30 | | Release date: | 2001-05-31 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a C-terminal coiled-coil domain from bovine IF(1): the inhibitor protein of F(1) ATPase.
J. Mol. Biol., 308, 2001
|
|
5M1S
 
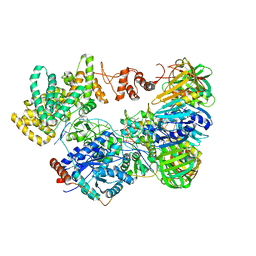 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | Descriptor: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2016-10-10 | | Release date: | 2017-01-18 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OLM
 
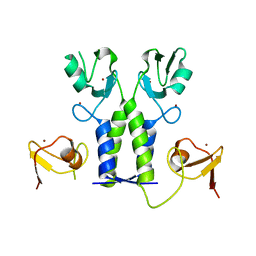 | | TRIM21 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, ZINC ION | | Authors: | James, L.C. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Intracellular antibody signalling is regulated by phosphorylation of the Fc receptor TRIM21.
Elife, 7, 2018
|
|
6QH7
 
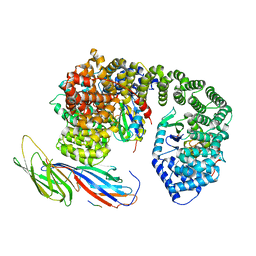 | | AP2 clathrin adaptor mu2T156-phosphorylated core with two cargo peptides in open+ conformation | | Descriptor: | ADAPTOR-RELATED PROTEIN COMPLEX 2, MU 2 SUBUNIT, C-TERMINAL DOMAIN, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6S53
 
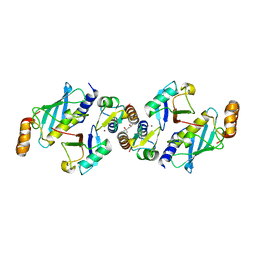 | | Crystal structure of TRIM21 RING domain in complex with an isopeptide-linked Ube2N~ubiquitin conjugate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, ... | | Authors: | Kiss, L, Boland, A, Neuhaus, D, James, L.C. | | Deposit date: | 2019-06-30 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases.
Nat Commun, 10, 2019
|
|
6TX3
 
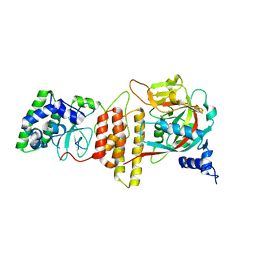 | | HPF1 bound to catalytic fragment of PARP2 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Histone PARylation factor 1, Poly [ADP-ribose] polymerase 2,Poly [ADP-ribose] polymerase 2 | | Authors: | Suskiewicz, M.J, Ahel, I. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation.
Nature, 579, 2020
|
|
8A58
 
 | |
2XTC
 
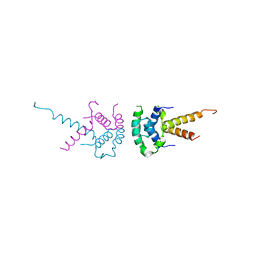 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2XTD
 
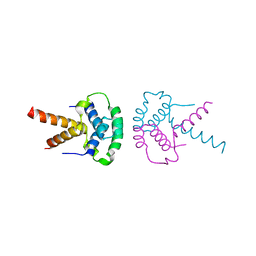 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | TBL1 F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2XTE
 
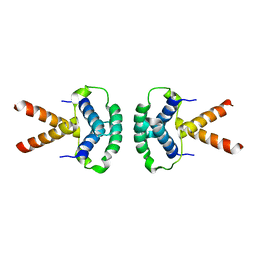 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2MY1
 
 | | Solution structure of Bud31p | | Descriptor: | Pre-mRNA-splicing factor BUD31, ZINC ION | | Authors: | van Roon, A.M, Yang, J, Mathieu, D, Bermel, W, Nagai, K, Neuhaus, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | (113) Cd NMR Experiments Reveal an Unusual Metal Cluster in the Solution Structure of the Yeast Splicing Protein Bud31p.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8UV0
 
 | |
1E6I
 
 | |
