5OPW
 
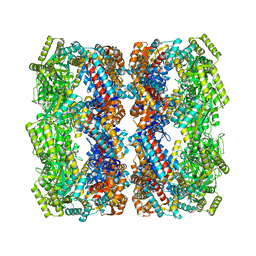 | | Crystal structure of the GroEL mutant A109C | | Descriptor: | 60 kDa chaperonin | | Authors: | Yan, X, Shi, Q, Bracher, A, Milicic, G, Singh, A.K, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | GroEL Ring Separation and Exchange in the Chaperonin Reaction.
Cell, 172, 2018
|
|
5OPX
 
 | | Crystal structure of the GroEL mutant A109C in complex with GroES and ADP BeF2 | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yan, X, Shi, Q, Bracher, A, Milicic, G, Singh, A.K, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | GroEL Ring Separation and Exchange in the Chaperonin Reaction.
Cell, 172, 2018
|
|
6XKC
 
 | | Crystal structure of E3 ligase | | Descriptor: | Protein fem-1 homolog C | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
7JYA
 
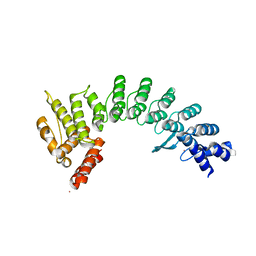 | | Crystal structure of E3 ligase in complex with peptide | | Descriptor: | ASN-ARG-ARG-ARG-ARG-TRP-ARG-GLU-ARG-GLN-ARG, Protein fem-1 homolog C, UNKNOWN ATOM OR ION | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-30 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
7EP0
 
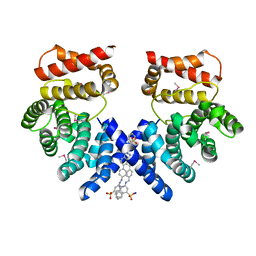 | | Crystal structure of ZYG11B bound to GSTE degron | | Descriptor: | Protein zyg-11 homolog B, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP2
 
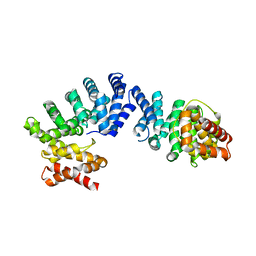 | | Crystal structure of ZYG11B bound to GGFN degron | | Descriptor: | Protein zyg-11 homolog B | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP3
 
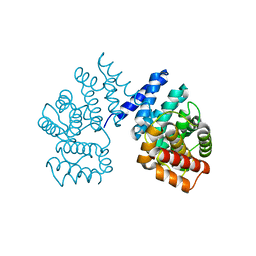 | | Crystal structure of ZER1 bound to GAGN degron | | Descriptor: | Protein zer-1 homolog | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.513 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP5
 
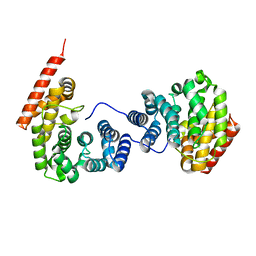 | | Crystal structure of ZER1 bound to GKLH degron | | Descriptor: | Protein zer-1 homolog | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP1
 
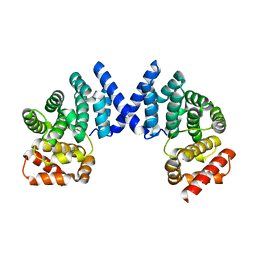 | | Crystal structure of ZYG11B bound to GFLH degron | | Descriptor: | Protein zyg-11 homolog B | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP4
 
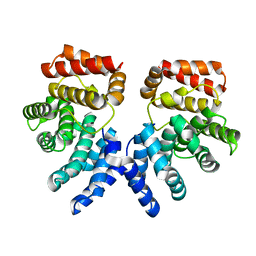 | | Crystal structure of ZER1 bound to GFLH degron | | Descriptor: | Protein zer-1 homolog | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
1BR6
 
 | | RICIN A CHAIN (RECOMBINANT) COMPLEX WITH PTEROIC ACID | | Descriptor: | PROTEIN (RICIN), PTEROIC ACID | | Authors: | Hollis, T, Yan, X, Svinth, M, Day, P, Monzingo, A.F, Milne, G.W.A, Robertus, J.D. | | Deposit date: | 1998-08-27 | | Release date: | 1998-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based identification of a ricin inhibitor.
J.Mol.Biol., 266, 1997
|
|
1BR5
 
 | | RICIN A CHAIN (RECOMBINANT) COMPLEX WITH NEOPTERIN | | Descriptor: | NEOPTERIN, PROTEIN (RICIN) | | Authors: | Day, P, Yan, X, Hollis, T, Svinth, M, Monzingo, A.F, Milne, G.W.A, Robertus, J.D. | | Deposit date: | 1998-08-26 | | Release date: | 1998-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based identification of a ricin inhibitor.
J.Mol.Biol., 266, 1997
|
|
4AIA
 
 | |
2IVY
 
 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG5
 
 | | CRYSTAL STRUCTURE OF A PUTATIVE PHOSPHOFRUCTOKINASE FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | FRUCTOSE 1-PHOSPHATE KINASE | | Authors: | Yan, X, Carter, L.G, Johnson, K.A, Liu, H, Dorward, M, McMahon, S.A, Oke, M, Powers, H, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2JG6
 
 | | CRYSTAL STRUCTURE OF A 3-METHYLADENINE DNA GLYCOSYLASE I FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | DNA-3-METHYLADENINE GLYCOSIDASE, ZINC ION | | Authors: | Yan, X, Carter, L.G, Liu, H, Dorward, M, McMahon, S.A, Johnson, K.A, Oke, M, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
3WZH
 
 | | Crystal structure of AfCsx3 | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein AF_1864 | | Authors: | Yuan, Y.A, Yan, X. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease.
Rna Biol., 12, 2015
|
|
3WZG
 
 | | Crystal structure of AfCsx3 | | Descriptor: | Uncharacterized protein AF_1864 | | Authors: | Yuan, Y.A, Yan, X. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease.
Rna Biol., 12, 2015
|
|
3WZI
 
 | | Crystal structure of AfCsx3 in complex with ssRNA | | Descriptor: | Uncharacterized protein AF_1864, ssRNA | | Authors: | Yuan, Y.A, Yan, X. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease.
Rna Biol., 12, 2015
|
|
8J4A
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtRPN10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors
Nat Commun, 15, 2024
|
|
8J49
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtSPL5 | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors
Nat Commun, 15, 2024
|
|
8J4B
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtSPL13 | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 13A, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors
Nat Commun, 15, 2024
|
|
8J48
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtGATA18 | | Descriptor: | GATA transcription factor 18, Sequence-variable mosaic (SVM) signal sequence domain-containing protein, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors
Nat Commun, 15, 2024
|
|
7XMW
 
 | | Crystal structure of anti-CRISPR protein AcrVIA2 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AcrVIA2, SELENIUM ATOM, ... | | Authors: | Yan, X, Li, X, Song, G. | | Deposit date: | 2022-04-27 | | Release date: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of AcrVIA2 and its binding mechanism to CRISPR-Cas13a.
Biochem.Biophys.Res.Commun., 612, 2022
|
|
6K3J
 
 | | Solution structure of APOBEC3G-CD2 with ssDNA, Product A | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*UP*(IUR)P*AP*AP*TP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Cao, C, Yan, X, Lan, W, Wang, C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Investigations on the Interactions between Cytidine Deaminase Human APOBEC3G and DNA.
Chem Asian J, 14, 2019
|
|
