1GLV
 
 | |
3LCK
 
 | |
2RT8
 
 | | Structure of metallo-dna in solution | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*CP*GP*CP*G)-3'), MERCURY (II) ION | | Authors: | Yamaguchi, H, Sebera, J, Kondo, J, Oda, S, Komuro, T, Kawamura, T, Dairaku, T, Kondo, Y, Okamoto, I, Ono, A, Burda, J.V, Kojima, C, Sychrovsky, V, Tanaka, Y. | | Deposit date: | 2013-06-18 | | Release date: | 2014-03-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The structure of metallo-DNA with consecutive thymine-HgII-thymine base pairs explains positive entropy for the metallo base pair formation.
Nucleic Acids Res., 42, 2014
|
|
8JPW
 
 | | Crystal Structure of Single-chain L-Glutamate Oxidase Mutant from Streptomyces sp. X-119-6 | | Descriptor: | 2-OXOGLUTARIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase, ... | | Authors: | Yamaguchi, H, Takahashi, K, Tatsumi, M, Tagami, U, Mizukoshi, T, Miyano, H, Sugiki, M. | | Deposit date: | 2023-06-13 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Development of a novel single-chain l-glutamate oxidase from Streptomyces sp. X-119-6 by inserting flexible linkers
Enzyme.Microb.Technol., 170, 2023
|
|
2F2U
 
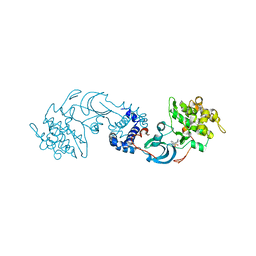 | | crystal structure of the Rho-kinase kinase domain | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, Rho-associated protein kinase 2 | | Authors: | Yamaguchi, H, Hakoshima, T. | | Deposit date: | 2005-11-18 | | Release date: | 2006-04-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism for the regulation of rho-kinase by dimerization and its inhibition by fasudil
Structure, 14, 2006
|
|
2H9V
 
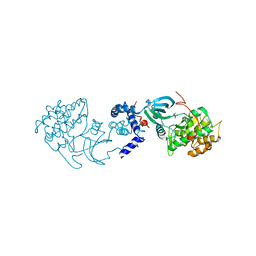 | | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, Rho-associated protein kinase 2 | | Authors: | Yamaguchi, H, Miwa, Y, Kasa, M, Kitano, K, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2006-06-12 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y-27632
J.Biochem.(Tokyo), 140, 2006
|
|
1IAJ
 
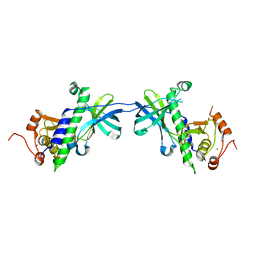 | | CRYSTAL STRUCTURE OF THE ATYPICAL PROTEIN KINASE DOMAIN OF A TRP CA-CHANNEL, CHAK (APO) | | Descriptor: | TRANSIENT RECEPTOR POTENTIAL-RELATED PROTEIN, ZINC ION | | Authors: | Yamaguchi, H, Matsushita, M, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity.
Mol.Cell, 7, 2001
|
|
1IAH
 
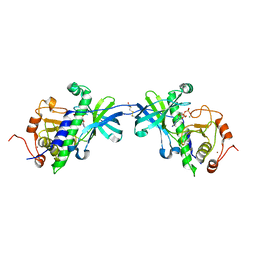 | | CRYSTAL STRUCTURE OF THE ATYPICAL PROTEIN KINASE DOMAIN OF A TRP CA-CHANNEL, CHAK (ADP-MG COMPLEX) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yamaguchi, H, Matsushita, M, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity.
Mol.Cell, 7, 2001
|
|
1IA9
 
 | | CRYSTAL STRUCTURE OF THE ATYPICAL PROTEIN KINASE DOMAIN OF A TRP CA-CHANNEL, CHAK (AMPPNP COMPLEX) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, TRANSIENT RECEPTOR POTENTIAL-RELATED PROTEIN, ... | | Authors: | Yamaguchi, H, Matsushita, M, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity.
Mol.Cell, 7, 2001
|
|
5ZBC
 
 | | Crystal structure of Se-Met tryptophan oxidase (C395A mutant) from Chromobacterium violaceum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA | | Authors: | Yamaguchi, H, Tatsumi, M, Takahashi, K, Tagami, U, Sugiki, M, Kashiwagi, T, Okazaki, S, Mizukoshi, T, Asano, Y. | | Deposit date: | 2018-02-11 | | Release date: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein engineering for improving the thermostability of tryptophan oxidase and insights from structural analysis.
J. Biochem., 164, 2018
|
|
5ZBD
 
 | | Crystal structure of tryptophan oxidase (C395A mutant) from Chromobacterium violaceum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, TRYPTOPHAN | | Authors: | Yamaguchi, H, Tatsumi, M, Takahashi, K, Tagami, U, Sugiki, M, Kashiwagi, T, Okazaki, S, Mizukoshi, T, Asano, Y. | | Deposit date: | 2018-02-11 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein engineering for improving the thermostability of tryptophan oxidase and insights from structural analysis.
J. Biochem., 164, 2018
|
|
6ACF
 
 | | structure of leucine dehydrogenase from Geobacillus stearothermophilus by cryo-EM | | Descriptor: | Leucine dehydrogenase | | Authors: | Yamaguchi, H, Kamegawa, A, Nakata, K, Kashiwagi, T, Mizukoshi, T, Fujiyoshi, Y, Tani, K. | | Deposit date: | 2018-07-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy
J. Struct. Biol., 205, 2019
|
|
6ACH
 
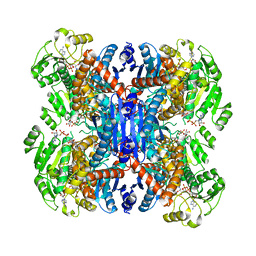 | | Structure of NAD+-bound leucine dehydrogenase from Geobacillus stearothermophilus by cryo-EM | | Descriptor: | Leucine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamaguchi, H, Kamegawa, A, Nakata, K, Kashiwagi, T, Mizukoshi, T, Fujiyoshi, Y, Tani, K. | | Deposit date: | 2018-07-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy
J. Struct. Biol., 205, 2019
|
|
1X1D
 
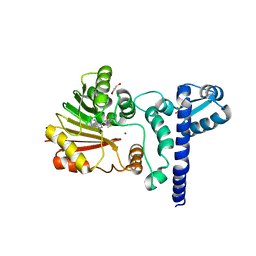 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine and Zn-bacteriopheophorbide d | | Descriptor: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-04 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1X1B
 
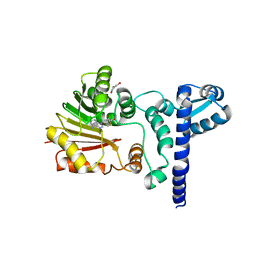 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine | | Descriptor: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-03 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1X19
 
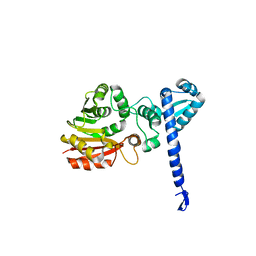 | | Crystal structure of BchU involved in bacteriochlorophyll c biosynthesis | | Descriptor: | CrtF-related protein, SULFATE ION | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-02 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1X1C
 
 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine and Zn2+ | | Descriptor: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-03 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1X1A
 
 | | Crystal structure of BchU complexed with S-adenosyl-L-methionine | | Descriptor: | CrtF-related protein, GLYCEROL, S-ADENOSYLMETHIONINE, ... | | Authors: | Yamaguchi, H, Wada, K, Fukuyama, K. | | Deposit date: | 2005-04-03 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1YSE
 
 | | Solution structure of the MAR-binding domain of SATB1 | | Descriptor: | DNA-binding protein SATB1 | | Authors: | Yamasaki, K, Yamaguchi, H. | | Deposit date: | 2005-02-08 | | Release date: | 2006-01-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and DNA-binding Mode of the Matrix Attachment Region-binding Domain of the Transcription Factor SATB1 That Regulates the T-cell Maturation
J.Biol.Chem., 281, 2006
|
|
1GSH
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 7.5 | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Kato, H, Yamaguchi, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
1AVK
 
 | | CRYSTAL STRUCTURES OF THE COPPER-CONTAINING AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS IN THE HOLO-AND APO-FORMS: IMPLICATIONS FOR THE BIOGENESIS OF TOPA QUINONE | | Descriptor: | AMINE OXIDASE | | Authors: | Wilce, M.C.J, Guss, J.M, Freeman, H.C, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 1997-09-16 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the copper-containing amine oxidase from Arthrobacter globiformis in the holo and apo forms: implications for the biogenesis of topaquinone.
Biochemistry, 36, 1997
|
|
8X36
 
 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form B) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8X35
 
 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP, IPP, and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
8X37
 
 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
2GLT
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
