5Y9N
 
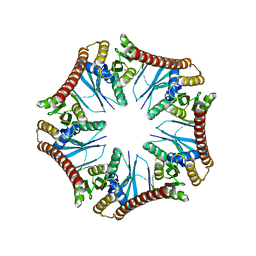 | | Crystal structure of Pyrococcus furiosus PbaA (monoclinic form), an archaeal homolog of proteasome-assembly chaperone | | Descriptor: | CHLORIDE ION, PbaA | | Authors: | Yagi-Utsumi, M, Sikdar, A, Kozai, T, Inoue, R, Sugiyama, M, Uchihashi, T, Satoh, T, Kato, K. | | Deposit date: | 2017-08-26 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Conversion of functionally undefined homopentameric protein PbaA into a proteasome activator by mutational modification of its C-terminal segment conformation
Protein Eng. Des. Sel., 31, 2018
|
|
5CRW
 
 | |
7Y8Q
 
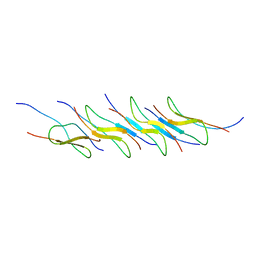 | | Amyloid-beta assemblage on GM1-containing membranes | | Descriptor: | Amyloid-beta protein 40 | | Authors: | Yagi-Utsumi, M, Itoh, S.G, Okumura, H, Yanagisawa, K, Kato, K, Nishimura, K. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-20 | | Method: | SOLID-STATE NMR | | Cite: | The Double-Layered Structure of Amyloid-beta Assemblage on GM1-Containing Membranes Catalytically Promotes Fibrillization.
Acs Chem Neurosci, 14, 2023
|
|
8IC9
 
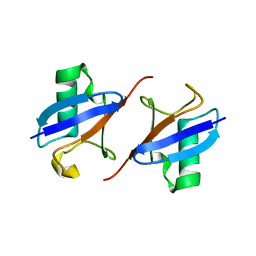 | | Lys48-linked K48C-diubiquitin | | Descriptor: | Polyubiquitin-B, Ubiquitin | | Authors: | Hiranyakorn, M, Yagi-Utsumi, M, Yanaka, S, Ohtsuka, N, Momiyama, N, Satoh, T, Kato, K. | | Deposit date: | 2023-02-11 | | Release date: | 2023-04-26 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mutational and Environmental Effects on the Dynamic Conformational Distributions of Lys48-Linked Ubiquitin Chains.
Int J Mol Sci, 24, 2023
|
|
6JPT
 
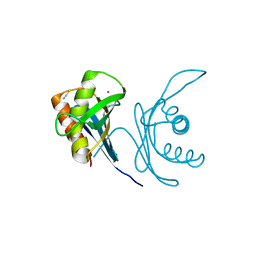 | | Crystal structure of human PAC3 homodimer (trigonal form) | | Descriptor: | POTASSIUM ION, Proteasome assembly chaperone 3, THIOCYANATE ION | | Authors: | Satoh, T, Yagi-Utsumi, M, Okamoto, K, Kurimoto, E, Tanaka, K, Kato, K. | | Deposit date: | 2019-03-27 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Molecular and Structural Basis of the Proteasome alpha Subunit Assembly Mechanism Mediated by the Proteasome-Assembling Chaperone PAC3-PAC4 Heterodimer.
Int J Mol Sci, 20, 2019
|
|
4YGE
 
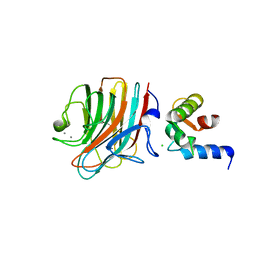 | | Crystal structure of ERGIC-53/MCFD2, trigonal calcium-bound form 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4YGB
 
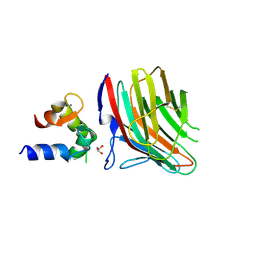 | | Crystal structure of ERGIC-53/MCFD2, monoclinic calcium-free form | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4YGC
 
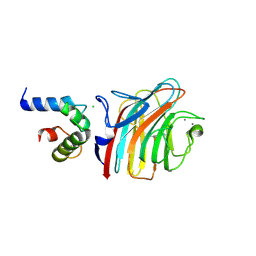 | | Crystal structure of ERGIC-53/MCFD2, monoclinic calcium-bound form 1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4YGD
 
 | | Crystal structure of ERGIC-53/MCFD2, monoclinic calcium-bound form 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Nishio, M, Yagi-Utsumi, M, Suzuki, K, Anzai, T, Mizushima, T, Kamiya, Y, Kato, K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystallographic snapshots of the EF-hand protein MCFD2 complexed with the intracellular lectin ERGIC-53 involved in glycoprotein transport.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3A33
 
 | | UbcH5b~Ubiquitin Conjugate | | Descriptor: | GLYCEROL, Ubiquitin, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Sakata, E, Satoh, T, Yamamoto, S, Yamaguchi, Y, Yagi-Utsumi, M, Kurimoto, E, Wakatsuki, S, Kato, K. | | Deposit date: | 2009-06-08 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of UbcH5b~Ubiquitin Intermediate: Insight into the Formation of the Self-Assembled E2~Ub Conjugates
Structure, 18, 2010
|
|
2RSU
 
 | | Alternative structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Kitazawa, S, Kameda, T, Yagi-Utsumi, M, Kato, K, Kitahara, R. | | Deposit date: | 2012-06-15 | | Release date: | 2013-03-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Q41N Variant of Ubiquitin as a Model for the Alternatively Folded N2 State of Ubiquitin
Biochemistry, 52, 2013
|
|
3WHJ
 
 | | Crystal structure of Nas2 N-terminal domain | | Descriptor: | CADMIUM ION, Probable 26S proteasome regulatory subunit p27, SULFATE ION | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHK
 
 | | Crystal structure of PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Proteasome-activating nucleotidase, 26S protease regulatory subunit 6A | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHL
 
 | | Crystal structure of Nas2 N-terminal domain complexed with PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Probable 26S proteasome regulatory subunit p27, Proteasome-activating nucleotidase, ... | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
7CAP
 
 | | Cyclic Lys48-linked triubiquitin | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Hiranyakorn, M, Yanaka, S, Satoh, T, Wilasri, T, Jityuti, B, Yagi-Utsumi, T, Kato, K. | | Deposit date: | 2020-06-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | NMR Characterization of Conformational Interconversions of Lys48-Linked Ubiquitin Chains.
Int J Mol Sci, 21, 2020
|
|
4DBG
 
 | | Crystal structure of HOIL-1L-UBL complexed with a HOIP-UBA derivative | | Descriptor: | RING finger protein 31, RanBP-type and C3HC4-type zinc finger-containing protein 1 | | Authors: | Yagi, H, Hiromoto, T, Mizushima, T, Kurimoto, E, Kato, K. | | Deposit date: | 2012-01-15 | | Release date: | 2012-04-04 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A non-canonical UBA-UBL interaction forms the linear-ubiquitin-chain assembly complex
Embo Rep., 13, 2012
|
|
2KP1
 
 | |
2KP2
 
 | |
5WTQ
 
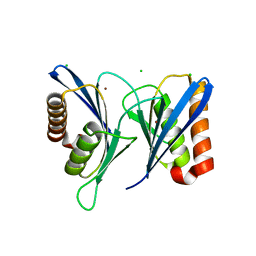 | | Crystal structure of human proteasome-assembling chaperone PAC4 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Proteasome assembly chaperone 4 | | Authors: | Kurimoto, E, Satoh, T, Ito, Y, Ishihara, E, Tanaka, K, Kato, K. | | Deposit date: | 2016-12-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human proteasome assembly chaperone PAC4 involved in proteasome formation
Protein Sci., 26, 2017
|
|
3AUL
 
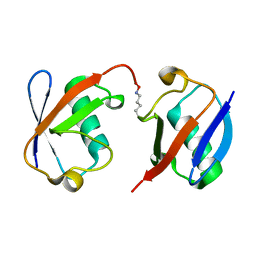 | | Crystal structure of wild-type Lys48-linked diubiquitin in an open conformation | | Descriptor: | Polyubiquitin-C | | Authors: | Hirano, T, Olivier, S, Yagi, M, Takemoto, E, Hiromoto, T, Satoh, T, Mizushima, T, Kato, K. | | Deposit date: | 2011-02-09 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Conformational dynamics of wild-type Lys48-linked diubiquitin in solution
J.Biol.Chem., 286, 2011
|
|
2DOS
 
 | |
2RU6
 
 | | The pure alternative state of ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Kitazawa, S, Kameda, T, Kumo, A, Utsumi, M, Baxter, N, Kato, K, Williamson, M.P, Kitahara, R. | | Deposit date: | 2013-12-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Close Identity between Alternatively Folded State N2 of Ubiquitin and the Conformation of the Protein Bound to the Ubiquitin-Activating Enzyme
Biochemistry, 53, 2014
|
|
3WSY
 
 | | SorLA Vps10p domain in complex with its own propeptide fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin-related receptor, peptide from Sortilin-related receptor | | Authors: | Kitago, Y, Nakata, Z, Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2014-03-30 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for amyloidogenic peptide recognition by sorLA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3WSZ
 
 | | SorLA Vps10p domain in complex with Abeta-derived peptide | | Descriptor: | 10-mer peptide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin-related receptor | | Authors: | Kitago, Y, Nakata, Z, Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2014-03-30 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis for amyloidogenic peptide recognition by sorLA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3WSX
 
 | | SorLA Vps10p domain in ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kitago, Y, Nakata, Z, Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2014-03-30 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for amyloidogenic peptide recognition by sorLA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
