8IZJ
 
 | |
6IGR
 
 | | Crystal structure of S9 peptidase (S514A mutant in inactive state) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Gaur, N.K, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IGP
 
 | | Crystal structure of S9 peptidase (inactive state)from Deinococcus radiodurans R1 in P212121 | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IGQ
 
 | | Crystal structure of inactive state of S9 peptidase from Deinococcus radiodurans R1 (PMSF treated) | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL, ... | | Authors: | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IKG
 
 | | Crystal structure of substrate-bound S9 peptidase (S514A mutant) from Deinococcus radiodurans | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL, ... | | Authors: | Yadav, P, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IRU
 
 | |
5YZO
 
 | | Crystal structure of S9 peptidase mutant (S514A) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative, DIMETHYL SULFOXIDE, ... | | Authors: | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
5YZN
 
 | | Crystal structure of S9 peptidase (active form) from Deinococcus radiodurans R1 | | Descriptor: | Acyl-peptide hydrolase, putative | | Authors: | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
5YZM
 
 | | Crystal structure of S9 peptidase (inactive form) from Deinococcus radiodurans R1 | | Descriptor: | ACETATE ION, Acyl-peptide hydrolase, putative | | Authors: | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6A4R
 
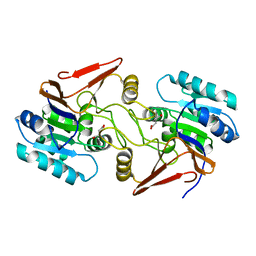 | | Crystal structure of aspartate bound peptidase E from Salmonella enterica | | Descriptor: | ASPARTIC ACID, Peptidase E | | Authors: | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6A4S
 
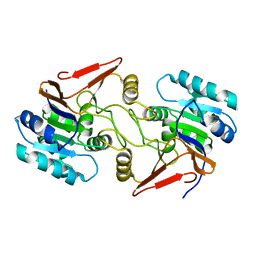 | | Crystal structure of peptidase E with ordered active site loop from Salmonella enterica | | Descriptor: | Peptidase E | | Authors: | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6A4T
 
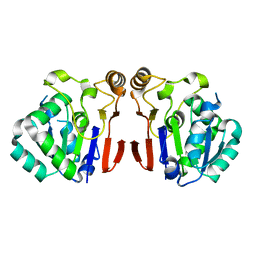 | | Crystal structure of Peptidase E from Deinococcus radiodurans R1 | | Descriptor: | Peptidase E | | Authors: | Yadav, P, Goyal, V.G, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic triad heterogeneity in S51 peptidase family: Structural basis for functional variability.
Proteins, 87, 2019
|
|
7XI2
 
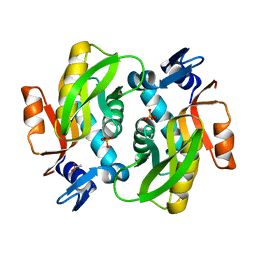 | |
7Y94
 
 | |
1LXY
 
 | | Crystal Structure of Arginine Deiminase covalently linked with L-citrulline | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine Deiminase, CITRULLINE | | Authors: | Das, K, Buttler, G.H, Kwiatkowski, V, Yadav, P, Arnold, E. | | Deposit date: | 2002-06-06 | | Release date: | 2004-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of arginine deiminase with covalent reaction intermediates; implications for catalytic mechanism
Structure, 12, 2004
|
|
5GIV
 
 | | Crystal structure of M32 carboxypeptidase from Deinococcus radiodurans R1 | | Descriptor: | ACETATE ION, Carboxypeptidase 1, ZINC ION | | Authors: | Sharma, B, Singh, R, Yadav, P, Ghosh, B, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active site gate of M32 carboxypeptidases illuminated by crystal structure and molecular dynamics simulations
Biochim. Biophys. Acta, 1865, 2017
|
|
1S9R
 
 | | CRYSTAL STRUCTURE OF ARGININE DEIMINASE COVALENTLY LINKED WITH A REACTION INTERMEDIATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ARGININE, Arginine deiminase, ... | | Authors: | Das, K, Buttler, G.H, Kwiatkowski, V, Clark Jr, A.D, Yadav, P, Arnold, E. | | Deposit date: | 2004-02-05 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Arginine Deiminase with Covalent Reaction
Intermediates: Implications for Catalytic Mechanism
Structure, 12, 2004
|
|
8WT1
 
 | | Crystal structure of S9 carboxypeptidase from Geobacillus sterothermophilus | | Descriptor: | ALANINE, CITRATE ANION, GLYCEROL, ... | | Authors: | Chandravanshi, K, Kumar, A, Sen, C, Singh, R, Bhange, G.B, Makde, R.D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
Febs Lett., 598, 2024
|
|
