6XR7
 
 | |
6XR6
 
 | |
6XRG
 
 | |
4R12
 
 | | Crystal structure of the gamma-secretase component Nicastrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xie, T, Yan, C, Zhou, R, Zhao, Y, Sun, L, Yang, G, Lu, P, Ma, D, Shi, Y. | | Deposit date: | 2014-08-03 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the gamma-secretase component nicastrin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WMS
 
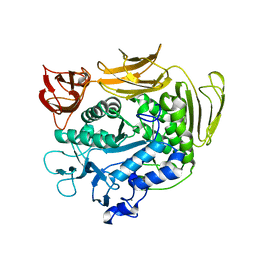 | | The crystal structure of Y195I mutant alpha-cyclodextrin glycosyltransferase from Paenibacillus macerans | | Descriptor: | Alpha-cyclodextrin glucanotransferase, CALCIUM ION | | Authors: | Xie, T, Hou, Y.J, Li, D.F, Yue, Y, Qian, S.J, Chao, Y.P. | | Deposit date: | 2013-11-24 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of a mutant Y195I alpha-cyclodextrin glycosyltransferase with switched product specificity from alpha-cyclodextrin to beta-/ gamma-cyclodextrin
J.Biotechnol., 182-183, 2014
|
|
4M68
 
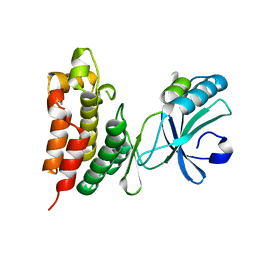 | | Crystal structure of the mouse MLKL kinase-like domain | | Descriptor: | GLYCEROL, Mixed lineage kinase domain-like protein | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
4M66
 
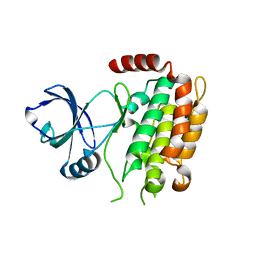 | | Crystal structure of the mouse RIP3 kinase domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
4M67
 
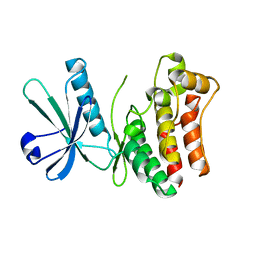 | | Crystal structure of the human MLKL kinase-like domain | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
4M69
 
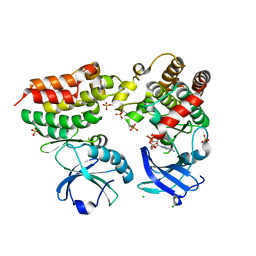 | | Crystal structure of the mouse RIP3-MLKL complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mixed lineage kinase domain-like protein, ... | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
4Q89
 
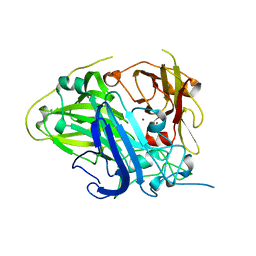 | |
4Q8B
 
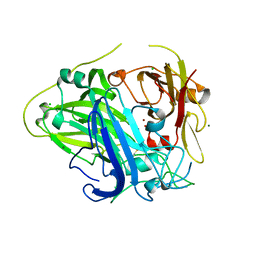 | |
8IAM
 
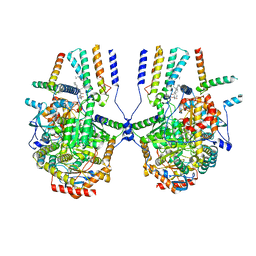 | | Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3D) complex | | Descriptor: | Chimera of Long chain base biosynthesis protein 1 and Serine palmitoyltransferase 1, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Xie, T, Gong, X. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Collaborative regulation of yeast SPT-Orm2 complex by phosphorylation and ceramide.
Cell Rep, 43, 2024
|
|
8IAK
 
 | |
8IAJ
 
 | | Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A) complex | | Descriptor: | N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, PYRIDOXAL-5'-PHOSPHATE, Protein ORM2, ... | | Authors: | Xie, T, Gong, X. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Collaborative regulation of yeast SPT-Orm2 complex by phosphorylation and ceramide.
Cell Rep, 43, 2024
|
|
8IZD
 
 | | Cryo-EM structure of the C26-CoA-bound Lac1-Lip1 complex | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Ceramide synthase LAC1, Ceramide synthase subunit LIP1, ... | | Authors: | Xie, T, Fang, Q, Gong, X. | | Deposit date: | 2023-04-07 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure and mechanism of a eukaryotic ceramide synthase complex.
Embo J., 42, 2023
|
|
8IZF
 
 | | Cryo-EM structure of the Lac1-Lip1 (Lip1-S74F) complex | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Ceramide synthase LAC1, Ceramide synthase subunit LIP1 | | Authors: | Xie, T, Fang, Q, Gong, X. | | Deposit date: | 2023-04-07 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure and mechanism of a eukaryotic ceramide synthase complex.
Embo J., 42, 2023
|
|
8S9P
 
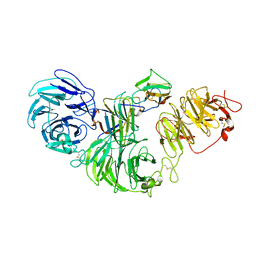 | | 1:1:1 agrin/LRP4/MuSK complex | | Descriptor: | Agrin, Low-density lipoprotein receptor-related protein 4, Muscle, ... | | Authors: | Xie, T, Xu, G.J, Liu, Y, Quade, B, Lin, W.C, Bai, X.C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-05-17 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the assembly of the agrin/LRP4/MuSK signaling complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2LKM
 
 | |
7W02
 
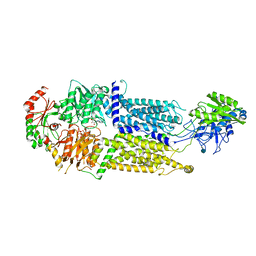 | | Cryo-EM structure of ATP-bound ABCA3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xie, T, Zhang, Z.K, Yue, J, Gong, X. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human surfactant lipid transporter ABCA3.
Sci Adv, 8, 2022
|
|
7W01
 
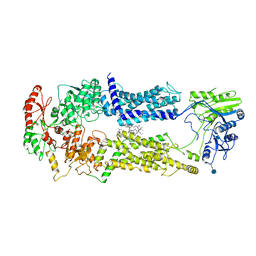 | | Cryo-EM structure of nucleotide-free ABCA3 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xie, T, Zhang, Z.K, Yue, J, Gong, X. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human surfactant lipid transporter ABCA3.
Sci Adv, 8, 2022
|
|
3UDB
 
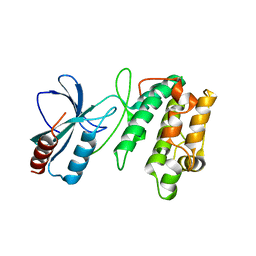 | | Crystal structure of SnRK2.6 | | Descriptor: | CHLORIDE ION, Serine/threonine-protein kinase SRK2E | | Authors: | Xie, T, Ren, R, Pang, Y, Yan, C. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.567 Å) | | Cite: | Molecular mechanism for the inhibition of a critical component in the Arabidopsis thaliana abscisic acid signal transduction pathways, SnRK2.6, by the protein phosphatase ABI1
to be published
|
|
4ITJ
 
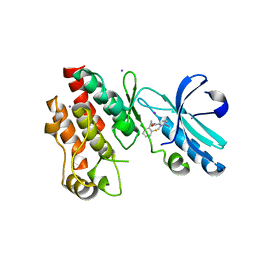 | | Crystal structure of RIP1 kinase in complex with necrostatin-4 | | Descriptor: | IODIDE ION, N-[(1S)-1-(2-chloro-6-fluorophenyl)ethyl]-5-cyano-1-methyl-1H-pyrrole-2-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
4ITI
 
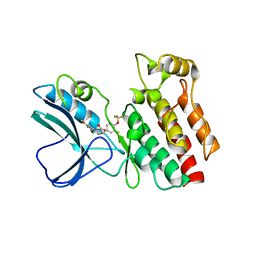 | | Crystal structure of RIP1 kinase in complex with necrostatin-3 analog | | Descriptor: | 1-{(3S,3aS)-3-[3-fluoro-4-(trifluoromethoxy)phenyl]-8-methoxy-3,3a,4,5-tetrahydro-2H-benzo[g]indazol-2-yl}-2-hydroxyethanone, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
4ITH
 
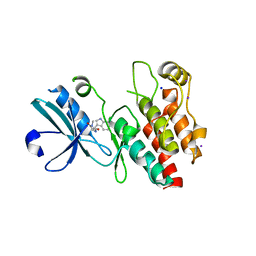 | | Crystal structure of RIP1 kinase in complex with necrostatin-1 analog | | Descriptor: | (5R)-5-[(7-chloro-1H-indol-3-yl)methyl]-3-methylimidazolidine-2,4-dione, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1, ... | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
2LD7
 
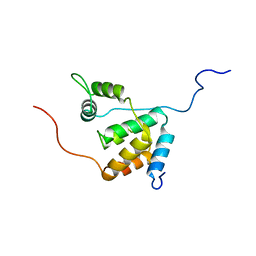 | | Solution structure of the mSin3A PAH3-SAP30 SID complex | | Descriptor: | Histone deacetylase complex subunit SAP30, Paired amphipathic helix protein Sin3a | | Authors: | Xie, T, He, Y, Korkeamaki, H, Zhang, Y, Imhoff, R, Lohi, O, Radhakrishnan, I. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the 30-kDa Sin3-associated protein (SAP30) in complex with the mammalian Sin3A corepressor and its role in nucleic acid binding.
J.Biol.Chem., 286, 2011
|
|
