1B87
 
 | |
6XAR
 
 | |
4M4Z
 
 | |
4PDP
 
 | |
1JPA
 
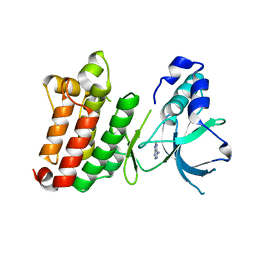 | | Crystal Structure of unphosphorylated EphB2 receptor tyrosine kinase and juxtamembrane region | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, neural kinase, Nuk=Eph/Elk/Eck family receptor-like tyrosine kinase | | Authors: | Wybenga-Groot, L.E, Pawson, T, Sicheri, F. | | Deposit date: | 2001-08-01 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region.
Cell(Cambridge,Mass.), 106, 2001
|
|
4TZI
 
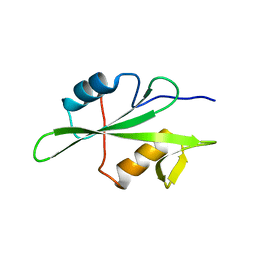 | | Structure of unliganded Lyn SH2 domain | | Descriptor: | Tyrosine-protein kinase Lyn | | Authors: | Wybenga-Groot, L.E. | | Deposit date: | 2014-07-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tyrosine Phosphorylation of the Lyn Src Homology 2 (SH2) Domain Modulates Its Binding Affinity and Specificity.
Mol.Cell Proteomics, 14, 2015
|
|
4PDS
 
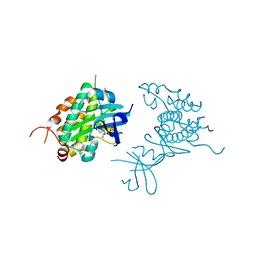 | | Crystal structure of Rad53 kinase domain and SCD2 in complex with AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase RAD53 | | Authors: | Ho, C.S, Wybenga-Groot, L.E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2014-04-21 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of Rad53 kinase activation by dimerization and activation segment exchange.
Cell Signal., 26, 2014
|
|
2HEN
 
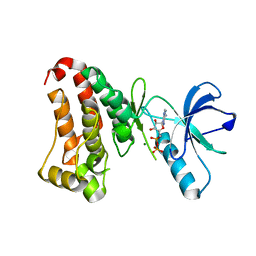 | |
2HEL
 
 | |
1N71
 
 | | Crystal structure of aminoglycoside 6'-acetyltransferase type Ii in complex with coenzyme A | | Descriptor: | COENZYME A, SULFATE ION, aac(6')-Ii | | Authors: | Burk, D.L, Ghuman, N, Wybenga-Groot, L.E, Berghuis, A.M. | | Deposit date: | 2002-11-12 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the AAC(6')-Ii antibiotic resistance enzyme at 1.8 A resolution; examination of oligomeric arrangements in GNAT superfamily members
Protein Sci., 12, 2003
|
|
2H6D
 
 | | Protein Kinase Domain of the Human 5'-AMP-activated protein kinase catalytic subunit alpha-2 (AMPK alpha-2 chain) | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | | Authors: | Littler, D.R, Walker, J.R, Wybenga-Groot, L, Newman, E.M, Butler-Cole, C, Mackenzie, F, Finerty, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-31 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A conserved mechanism of autoinhibition for the AMPK kinase domain: ATP-binding site and catalytic loop refolding as a means of regulation.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2A4N
 
 | | Crystal structure of aminoglycoside 6'-N-acetyltransferase complexed with coenzyme A | | Descriptor: | COENZYME A, SULFATE ION, aac(6')-Ii | | Authors: | Burk, D.L, Xiong, B, Breitbach, C, Berghuis, A.M. | | Deposit date: | 2005-06-29 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of aminoglycoside acetyltransferase AAC(6')-Ii in a novel crystal form: structural and normal-mode analyses.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
