2KXJ
 
 | | Solution structure of UBX domain of human UBXD2 protein | | Descriptor: | UBX domain-containing protein 4 | | Authors: | Wu, Q, Huang, H, Zhang, J, Hu, Q, Wu, J, Shi, Y. | | Deposit date: | 2010-05-06 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution strcture of UBX domain of human UBXD2 protein
To be Published
|
|
1YZ3
 
 | | Structure of human pnmt complexed with cofactor product adohcy and inhibitor SK&F 64139 | | Descriptor: | 7,8-DICHLORO-1,2,3,4-TETRAHYDROISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, Q, Gee, C.L, Lin, F, Martin, J.L, Grunewald, G.L, McLeish, M.J. | | Deposit date: | 2005-02-27 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, mutagenic, and kinetic analysis of the binding of substrates and inhibitors of human phenylethanolamine N-methyltransferase
J.Med.Chem., 48, 2005
|
|
4Y18
 
 | |
4Y2G
 
 | |
2KB2
 
 | | BlrP1 BLUF | | Descriptor: | BlrP1, FLAVIN MONONUCLEOTIDE | | Authors: | Wu, Q, Gardner, K.H. | | Deposit date: | 2008-11-19 | | Release date: | 2009-04-07 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structure and insight into blue light-induced changes in the BlrP1 BLUF domain
Biochemistry, 48, 2009
|
|
3W03
 
 | | XLF-XRCC4 complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Wu, Q, Ochi, T, Matak-Vinkovic, D, Robinson, C.V, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2012-10-17 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (8.492 Å) | | Cite: | Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions
Biochem.Soc.Trans., 39, 2011
|
|
4LX3
 
 | | Conserved Residues that Modulate Protein trans-Splicing of Npu DnaE Split Intein | | Descriptor: | DNA polymerase III, alpha subunit, Nucleic acid binding, ... | | Authors: | Wu, Q, Gao, Z, Wei, Y, Ma, G, Zheng, Y, Dong, Y, Liu, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-25 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved residues that modulate protein trans-splicing of Npu DnaE split intein.
Biochem.J., 461, 2014
|
|
6EBU
 
 | | Crystal structure of Aquifex aeolicus LpxE | | Descriptor: | LpxE, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Wu, Q, Wang, S, Zhou, P. | | Deposit date: | 2018-08-07 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | The Lipid A 1-Phosphatase, LpxE, Functionally Connects Multiple Layers of Bacterial Envelope Biogenesis.
Mbio, 10, 2019
|
|
4I8B
 
 | | Crystal Structure of Thioredoxin from Schistosoma Japonicum | | Descriptor: | Thioredoxin | | Authors: | Wu, Q, Peng, Y, Zhao, J, Li, X, Fan, X, Zhou, X, Chen, J, Luo, Z, Shi, D. | | Deposit date: | 2012-12-03 | | Release date: | 2013-12-04 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression, characterization and crystal structure of thioredoxin from Schistosoma japonicum.
Parasitology, 142, 2015
|
|
7TAD
 
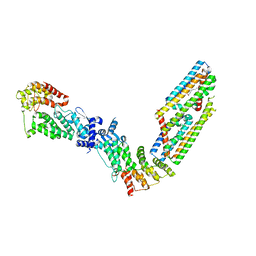 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
4WHG
 
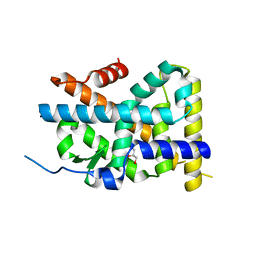 | | Crystal Structure of TR3 LBD in complex with Molecule 3 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)octan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Wang, Y, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4WHF
 
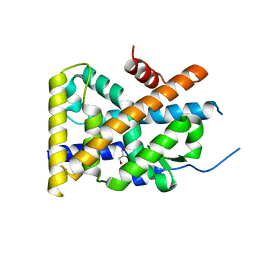 | | Crystal Structure of TR3 LBD in complex with 1-(3,4,5-trihydroxyphenyl)decan-1-one | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)decan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Wang, Y, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
7MK3
 
 | | Crystal structure of NPR1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Regulatory protein NPR1, ... | | Authors: | Cheng, J, Wu, Q, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
4RZF
 
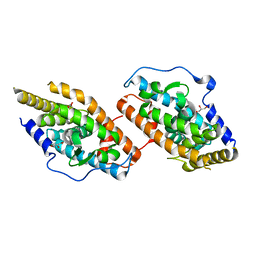 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, S441W mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
4RZG
 
 | | Crystal Structure Analysis of the DNPA-bounded NUR77 Ligand binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, pentyl (3,5-dihydroxy-2-nonanoylphenyl)acetate | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
7OR5
 
 | | Ternary complex of 14-3-3 sigma, NotchpS1917 phosphopeptide, and WQ162 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Neurogenic locus notch homolog protein 4, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-04 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
7OR7
 
 | | Ternary complex of 14-3-3 sigma, NotchpS1917 phosphopeptide, and WQ178 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-04 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
7OR3
 
 | | Ternary complex of 14-3-3 sigma, NotchpS1917 phosphopeptide, and WQ136 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
7ORG
 
 | | Ternary complex of 14-3-3 sigma, p27pT198 phosphopeptide, and WQ162 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
7ORH
 
 | | Ternary complex of 14-3-3 sigma, p27pT198 phosphopeptide, and WQ178 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
7ORS
 
 | | Ternary complex of 14-3-3 sigma, p27pT198 phosphopeptide, and WQ147 | | Descriptor: | (4~{S})-~{N}-[(5-carbamimidoyl-3-phenyl-thiophen-2-yl)methyl]oxepane-4-carboxamide, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
7OR8
 
 | | Ternary complex of 14-3-3 sigma, p27pT198 phosphopeptide, and WQ136 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
7ORT
 
 | | Ternary complex of 14-3-3 sigma, p27pT198 phosphopeptide, and WQ176 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
2AN5
 
 | | Structure of human PNMT complexed with S-adenosyl-homocysteine and an inhibitor, trans-(1S,2S)-2-amino-1-tetralol | | Descriptor: | PHOSPHATE ION, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | Deposit date: | 2005-08-11 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
