1ZFS
 
 | |
1ZRI
 
 | |
2MWC
 
 | |
2N56
 
 | |
2K2F
 
 | |
2K7O
 
 | | Ca2+-S100B, refined with RDCs | | Descriptor: | CALCIUM ION, Protein S100-B | | Authors: | Wright, N.T, Inman, K.G, Levine, J.A, Weber, D.J. | | Deposit date: | 2008-08-17 | | Release date: | 2008-11-18 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Refinement of the solution structure and dynamic properties of Ca(2+)-bound rat S100B.
J.Biomol.Nmr, 42, 2008
|
|
2KBM
 
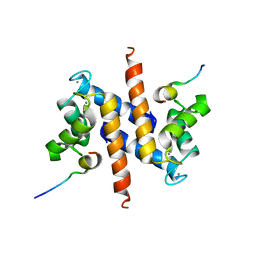 | | Ca-S100A1 interacting with TRTK12 | | Descriptor: | CALCIUM ION, F-actin-capping protein subunit alpha-2, Protein S100-A1 | | Authors: | Wright, N.T, Varney, K.M, Cannon, B.R, Morgan, M, Weber, D.J. | | Deposit date: | 2008-12-02 | | Release date: | 2009-02-10 | | Last modified: | 2018-08-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ca-S100A1-TRTK12
To be Published
|
|
2L8B
 
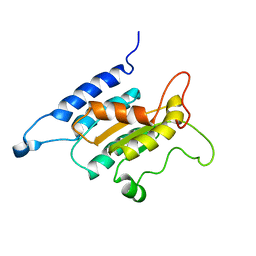 | | TraI (381-569) | | Descriptor: | Protein traI | | Authors: | Wright, N.T, Raththagala, M.U, Edwards, S, Krueger, S, Schildbach, J.F. | | Deposit date: | 2011-01-07 | | Release date: | 2012-01-11 | | Last modified: | 2023-02-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure and small angle scattering analysis of TraI (381-569).
Proteins, 80, 2012
|
|
6MG9
 
 | |
4RSV
 
 | |
6DL4
 
 | | Human Titin ZIg10 | | Descriptor: | Titin | | Authors: | Wright, N.T. | | Deposit date: | 2018-05-31 | | Release date: | 2019-08-21 | | Last modified: | 2020-03-04 | | Method: | SOLUTION NMR | | Cite: | Structural Insights on the Obscurin-Binding Domains in Titin.
Protein Pept.Lett., 25, 2018
|
|
5TZM
 
 | |
7R67
 
 | | Human obscurin Ig13 | | Descriptor: | Obscurin | | Authors: | Moncure, G.E, Wright, N.T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-03-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminus of obscurin is flexible in solution.
Proteins, 91, 2023
|
|
7R68
 
 | | Human obscurin Ig12 | | Descriptor: | Obscurin | | Authors: | Mauriello, G.E, Wright, N.T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-11-02 | | Last modified: | 2023-03-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminus of obscurin is flexible in solution.
Proteins, 91, 2023
|
|
3S3U
 
 | |
3TM2
 
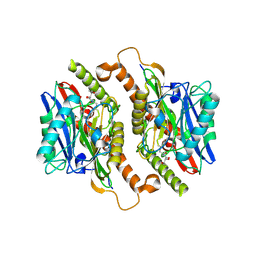 | | Crystal structure of mature ThnT with a covalently bound product mimic | | Descriptor: | (2R)-N-(4-chloro-3-oxobutyl)-2,4-dihydroxy-3,3-dimethylbutanamide, cysteine transferase | | Authors: | Schildbach, J.F, Wright, N.T, Buller, A.R. | | Deposit date: | 2011-08-30 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Autoproteolytic Activation of ThnT Results in Structural Reorganization Necessary for Substrate Binding and Catalysis.
J.Mol.Biol., 422, 2012
|
|
3TM1
 
 | |
2JUB
 
 | | Solution structure of IPI* | | Descriptor: | Internal protein I | | Authors: | Rifat, D, Wright, N.T, Varney, K.M, Weber, D.J, Black, L.W. | | Deposit date: | 2007-08-17 | | Release date: | 2007-12-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target.
J.Mol.Biol., 375, 2008
|
|
