6DE4
 
 | | Homo sapiens dihydrofolate reductase complexed with beta-NADPH and 3'-[(2R)-4-(2,4-diamino-6-ethylphenyl)but-3-yn-2-yl]-5'-methoxy-[1,1'-biphenyl]-4-carboxylic acid | | Descriptor: | 3'-[(2R)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-5'-methoxy[1,1'-biphenyl]-4-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hajian, B, Wright, D. | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
6DDS
 
 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-5-methoxy-phenyl]benzoic acid | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-5-methoxy-phenyl]benzoic acid, ACETATE ION, Dihydrofolate reductase, ... | | Authors: | Hajian, B, Wright, D, Scocchera, E. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
6DDW
 
 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid | | Descriptor: | Dihydrofolate reductase, GLYCEROL, N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid, ... | | Authors: | Hajian, B, Scocchera, E, Wright, D. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
6DDP
 
 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and 3'-[(2R)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-5'-methoxy[1,1'-biphenyl]-4-carboxylic acid | | Descriptor: | 3'-[(2R)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-5'-methoxy[1,1'-biphenyl]-4-carboxylic acid, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hajian, B, Wright, D. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
6DE5
 
 | |
6FAP
 
 | | Crystal structure of human BAZ2A PHD zinc finger in complex with Fr23 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2017-12-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6FKP
 
 | | Crystal structure of BAZ2A PHD zinc finger in complex with H3 10-mer AA mutant peptide | | Descriptor: | ALA-ARG-THR-ALA-ALA-THR-ALA-ARG, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6FI0
 
 | | Crystal structure of BAZ2A PHD zinc finger in complex with Fr 19 | | Descriptor: | 2-azanyl-1-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl)ethanone, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6FHQ
 
 | | Crystal structure of human BAZ2B PHD zinc finger in complex with Fr 21 | | Descriptor: | 2-azanyl-~{N}-(1,3-thiazol-2-yl)ethanamide, Bromodomain adjacent to zinc finger domain protein 2B, ZINC ION | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6FHU
 
 | | Crystal structure of BAZ2A PHD zinc finger in complex with H3 3-mer peptide | | Descriptor: | ALA-ARG-TAM, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-03-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6FI1
 
 | | Crystal structure of human BAZ2B PHD zinc finger in complex with Fr23 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, ZINC ION, ~{N}-(4-aminophenyl)-2-azanyl-ethanamide | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
7R6G
 
 | | Crystal structure of DfrA5 dihydrofolate reductase in complex with TRIMETHOPRIM and NADPH | | Descriptor: | Dihydrofolate reductase type 5, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Estrada, A, Wright, D, Krucinska, J, Erlandsen, H. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
6NPF
 
 | | Structure of E.coli enolase in complex with an analog of the natural product SF-2312 metabolite. | | Descriptor: | Enolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Erlandsen, H, Krucinska, J, Lombardo, M, Wright, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
7MYM
 
 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM and NADPH | | Descriptor: | ARGININE, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7MYL
 
 | |
7NAE
 
 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM | | Descriptor: | Dihydrofolate reductase, SULFATE ION, TRIMETHOPRIM | | Authors: | Estrada, A, Wright, D, Krucinska, J, Erlandsen, H. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
6I9I
 
 | |
6BFZ
 
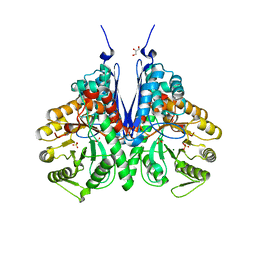 | | Crystal structure of enolase from E. coli with a mixture of apo form, substrate, and product form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PHOSPHOGLYCERIC ACID, Enolase, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
6BFY
 
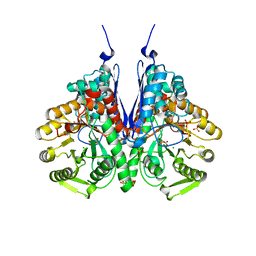 | | Crystal structure of enolase from Escherichia coli with bound 2-phosphoglycerate substrate | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Enolase, GLYCEROL, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
5LCJ
 
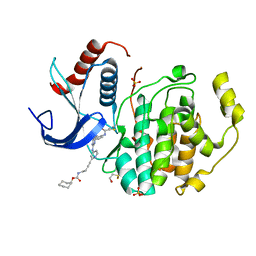 | | In-Gel Activity-Based Protein Profiling of a Clickable Covalent Erk 1/2 Inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, [(1~{R},4~{Z})-cyclooct-4-en-1-yl] ~{N}-[4-[4-[[5-chloranyl-4-[[2-(propanoylamino)phenyl]amino]pyrimidin-2-yl]amino]pyridin-2-yl]but-3-ynyl]carbamate | | Authors: | O'Reilly, M, Wright, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-20 | | Last modified: | 2016-08-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | In-gel activity-based protein profiling of a clickable covalent ERK1/2 inhibitor.
Mol Biosyst, 12, 2016
|
|
5LCK
 
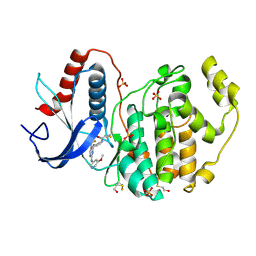 | | A Clickable Covalent ERK 1/2 Inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, ~{N}-[2-[[2-[(5-methoxypyridin-3-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]propanamide | | Authors: | O'Reilly, M, Wright, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-20 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In-gel activity-based protein profiling of a clickable covalent ERK1/2 inhibitor.
Mol Biosyst, 12, 2016
|
|
6D3Q
 
 | | Crystal structure of Escherichia coli enolase complexed with a natural inhibitor SF2312. | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Erlandsen, H, Krucinska, J, Hazeen, A, Wright, D. | | Deposit date: | 2018-04-16 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
