1XY2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF DEAMINO-OXYTOCIN. CONFORMATIONAL FLEXIBILITY AND RECEPTOR BINDING | | Descriptor: | OXYTOCIN | | Authors: | Cooper, S, Blundell, T.L, Pitts, J.E, Wood, S.P, Tickle, I.J. | | Deposit date: | 1987-06-05 | | Release date: | 1988-04-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding.
Science, 232, 1986
|
|
1XY1
 
 | | CRYSTAL STRUCTURE ANALYSIS OF DEAMINO-OXYTOCIN. CONFORMATIONAL FLEXIBILITY AND RECEPTOR BINDING | | Descriptor: | BETA-MERCAPTOPROPIONATE-OXYTOCIN | | Authors: | Husain, J, Blundell, T.L, Wood, S.P, Tickle, I.J, Cooper, S, Pitts, J.E. | | Deposit date: | 1987-06-05 | | Release date: | 1988-04-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding.
Science, 232, 1986
|
|
1B4E
 
 | | X-ray structure of 5-aminolevulinic acid dehydratase complexed with the inhibitor levulinic acid | | Descriptor: | GLYCEROL, LAEVULINIC ACID, PROTEIN (5-AMINOLEVULINIC ACID DEHYDRATASE), ... | | Authors: | Erskine, P.T, Cooper, J.B, Lewis, G, Spencer, P, Wood, S.P, Shoolingin-Jordan, P.M. | | Deposit date: | 1998-12-19 | | Release date: | 1999-12-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 5-aminolevulinic acid dehydratase from Escherichia coli complexed with the inhibitor levulinic acid at 2.0 A resolution.
Biochemistry, 38, 1999
|
|
1PPT
 
 | | X-RAY ANALYSIS (1.4-ANGSTROMS RESOLUTION) OF AVIAN PANCREATIC POLYPEPTIDE. SMALL GLOBULAR PROTEIN HORMONE | | Descriptor: | AVIAN PANCREATIC POLYPEPTIDE, ZINC ION | | Authors: | Blundell, T.L, Pitts, J.E, Tickle, I.J, Wood, S.P. | | Deposit date: | 1981-01-16 | | Release date: | 1981-02-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray analysis (1. 4-A resolution) of avian pancreatic polypeptide: Small globular protein hormone.
Proc.Natl.Acad.Sci.Usa, 78, 1981
|
|
4YP2
 
 | | Cleavage of nicotinamide adenine dinucleotides by the ribosome inactivating protein from Momordica charantia | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, Ribosome-inactivating protein momordin I | | Authors: | Vinkovic, M, Hussain, J, Wood, G.E, Gill, R, Wood, S.P. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Cleavage of nicotinamide adenine dinucleotide by the ribosome-inactivating protein from Momordica charantia.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1SAC
 
 | | THE STRUCTURE OF PENTAMERIC HUMAN SERUM AMYLOID P COMPONENT | | Descriptor: | ACETIC ACID, CALCIUM ION, SERUM AMYLOID P COMPONENT | | Authors: | White, H.E, Emsley, J, O'Hara, B.P, Oliva, G, Srinivasan, N, Tickle, I.J, Blundell, T.L, Pepys, M.B, Wood, S.P. | | Deposit date: | 1994-01-27 | | Release date: | 1994-05-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of pentameric human serum amyloid P component.
Nature, 367, 1994
|
|
1B09
 
 | |
1AW5
 
 | |
2IZP
 
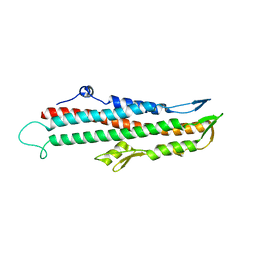 | | BipD - an invasion protein associated with the type-III secretion system of Burkholderia pseudomallei. | | Descriptor: | PUTATIVE MEMBRANE ANTIGEN | | Authors: | Erskine, P.T, Knight, M.J, Ruaux, A, Mikolajek, H, Wong-Fat-Sang, N, Withers, J, Gill, R, Wood, S.P, Wood, M, Fox, G.C, Cooper, J.B. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-06 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution Structure of Bipd: An Invasion Protein Associated with the Type III Secretion System of Burkholderia Pseudomallei.
J.Mol.Biol., 363, 2006
|
|
1P3H
 
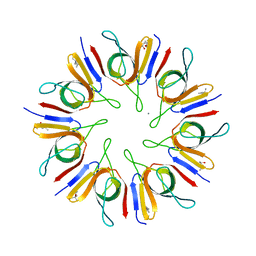 | | Crystal Structure of the Mycobacterium tuberculosis chaperonin 10 tetradecamer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 10 kDa chaperonin, CALCIUM ION | | Authors: | Roberts, M.M, Coker, A.R, Fossati, G, Mascagni, P, Coates, A.R.M, Wood, S.P, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops
J.BACTERIOL., 185, 2003
|
|
5JK4
 
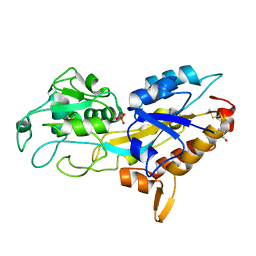 | | Phosphate-Binding Protein from Stenotrophomonas maltophilia. | | Descriptor: | Alkaline phosphatase, PHOSPHATE ION | | Authors: | Keegan, R, Waterman, D, Hopper, D, Coates, L, Guo, J, Coker, A.R, Erskine, P.T, Wood, S.P, Cooper, J.B. | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 angstrom resolution structure of a periplasmic phosphate-binding protein from Stenotrophomonas maltophilia: a crystallization contaminant identified by molecular replacement using the entire Protein Data Bank.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3EQ1
 
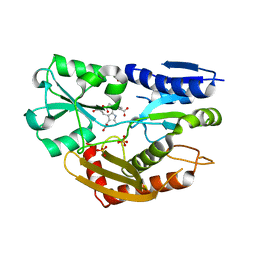 | | The Crystal Structure of Human Porphobilinogen Deaminase at 2.8A resolution | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase, SULFATE ION | | Authors: | Kolstoe, S.E, Gill, R, Mohammed, F, Wood, S.P. | | Deposit date: | 2008-09-30 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human porphobilinogen deaminase at 2.8 A: the molecular basis of acute intermittent porphyria
Biochem.J., 420, 2009
|
|
1U2E
 
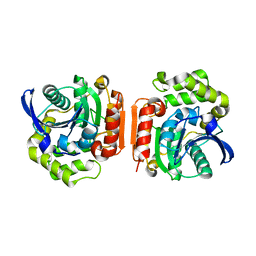 | | Crystal Structure of the C-C bond hydrolase MhpC | | Descriptor: | 2-hydroxy-6-ketonona-2,4-dienedioic acid hydrolase, CHLORIDE ION | | Authors: | Montgomery, M.G, Dunn, G, Mohammed, F, Robertson, T, Garcia, J.-L, Coker, A, Bugg, T.D.H, Wood, S.P. | | Deposit date: | 2004-07-19 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the C-C Bond Hydrolase MhpC Provides Insights into its Catalytic Mechanism
J.Mol.Biol., 346, 2005
|
|
1W6S
 
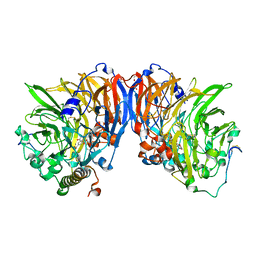 | | The high resolution structure of methanol dehydrogenase from methylobacterium extorquens | | Descriptor: | CALCIUM ION, GLYCEROL, METHANOL DEHYDROGENASE SUBUNIT 1, ... | | Authors: | Williams, P.A, Coates, L, Mohammed, F, Gill, R, Erskine, P.T, Wood, S.P, Anthony, C, Cooper, J.B. | | Deposit date: | 2004-08-23 | | Release date: | 2004-12-21 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Atomic Resolution Structure of Methanol Dehydrogenase from Methylobacterium Extorquens
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1W1Z
 
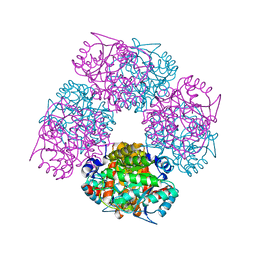 | | Structure of the plant like 5-Aminolaevulinic Acid Dehydratase from Chlorobium vibrioforme | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, LAEVULINIC ACID, MAGNESIUM ION | | Authors: | Coates, L, Beaven, G, Erskine, P.T, Beale, S.I, Avissar, Y.J, Gill, R, Mohammed, F, Wood, S.P, Shoolingin-Jordan, P, Cooper, J.B. | | Deposit date: | 2004-06-24 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the plant like 5-aminolaevulinic acid dehydratase from Chlorobium vibrioforme complexed with the inhibitor laevulinic acid at 2.6 A resolution.
J. Mol. Biol., 342, 2004
|
|
1W31
 
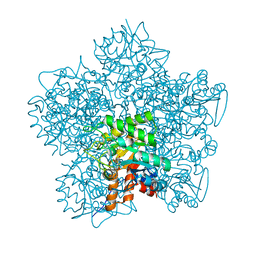 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 5-HYDROXYLAEVULINIC ACID COMPLEX | | Descriptor: | 5-HYDROXYLAEVULINIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Beaven, G.D.E, Gill, R, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2004-07-11 | | Release date: | 2005-08-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with the Inhibitor 5-Hydroxylaevulinic Acid
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5MDN
 
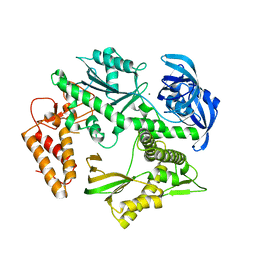 | | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | DNA polymerase, MAGNESIUM ION | | Authors: | Guo, J, Zhang, W, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Akhtar, M. | | Deposit date: | 2016-11-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5CF9
 
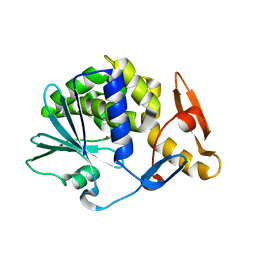 | | Cleavage of nicotinamide adenine dinucleotide by the ribosome inactivating protein of Momordica charantia - enzyme-NADP+ co-crystallisation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, Ribosome-inactivating protein momordin I | | Authors: | Vinkovic, M, Wood, S.P, Gill, R, Husain, J, Wood, G.E, Dunn, G. | | Deposit date: | 2015-07-08 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Cleavage of nicotinamide adenine dinucleotide by the ribosome-inactivating protein from Momordica charantia.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1IDS
 
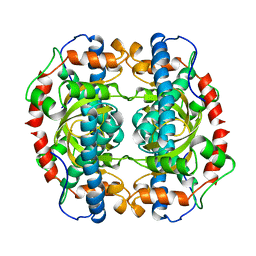 | | X-RAY STRUCTURE ANALYSIS OF THE IRON-DEPENDENT SUPEROXIDE DISMUTASE FROM MYCOBACTERIUM TUBERCULOSIS AT 2.0 ANGSTROMS RESOLUTIONS REVEALS NOVEL DIMER-DIMER INTERACTIONS | | Descriptor: | FE (III) ION, IRON SUPEROXIDE DISMUTASE | | Authors: | Cooper, J.B, Mcintyre, K, Wood, S.P, Zhang, Y, Young, D. | | Deposit date: | 1994-09-29 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of the iron-dependent superoxide dismutase from Mycobacterium tuberculosis at 2.0 Angstroms resolution reveals novel dimer-dimer interactions.
J.Mol.Biol., 246, 1995
|
|
1LGN
 
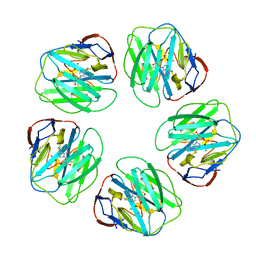 | |
2JXR
 
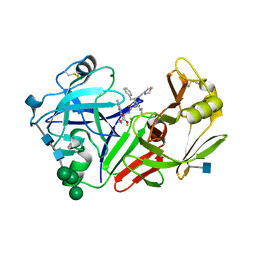 | | STRUCTURE OF YEAST PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide, PROTEINASE A, ... | | Authors: | Aguilar, C.F, Badasso, M, Dreyer, T, Cronin, N.B, Newman, M.P, Cooper, J.B, Hoover, D.J, Wood, S.P, Johnson, M.S, Blundell, T.L. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae.
J.Mol.Biol., 267, 1997
|
|
5OV5
 
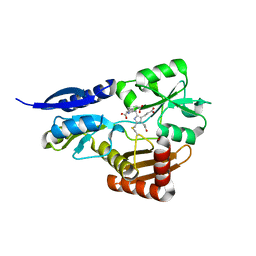 | | Bacillus megaterium porphobilinogen deaminase D82E mutant | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OT1
 
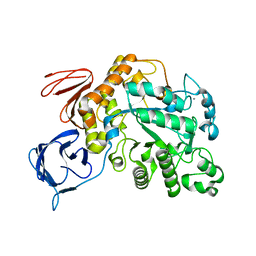 | | The type III pullulan hydrolase from Thermococcus kodakarensis | | Descriptor: | CALCIUM ION, Pullulanase type II, GH13 family | | Authors: | Guo, J, Coker, A.R, Wood, S.P, Cooper, J.B, Keegan, R, Ahmad, N, Muhammad, M.A, Rashid, N, Akhtar, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of the type III pullulan hydrolase from Thermococcus kodakarensis.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2W08
 
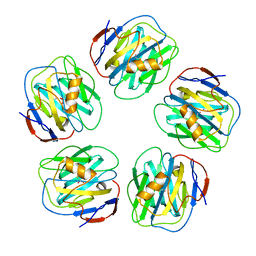 | | The structure of serum amyloid P component bound to 0-phospho- threonine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PHOSPHOTHREONINE, ... | | Authors: | Kolstoe, S.E, Pepys, M.B, Wood, S.P. | | Deposit date: | 2008-08-12 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Dissection of Alzheimer'S Disease Neuropathology by Depletion of Serum Amyloid P Component.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5OV6
 
 | | Bacillus megaterium porphobilinogen deaminase D82N mutant | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-2,5-dimethyl-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
