4YH2
 
 | |
3MAK
 
 | |
3F6F
 
 | |
3F63
 
 | |
3F6D
 
 | | Crystal Structure of a Genetically Modified Delta Class GST (adGSTD4-4) from Anopheles dirus, F123A, in Complex with S-Hexyl Glutathione | | Descriptor: | Glutathione transferase GST1-4, S-HEXYLGLUTATHIONE | | Authors: | Wongsantichon, J, Robinson, R.C, Ketterman, A.J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural contributions of delta class glutathione transferase active-site residues to catalysis
Biochem.J., 428, 2010
|
|
3G7I
 
 | |
3G7J
 
 | | Crystal Structure of a Genetically Modified Delta Class GST (adGSTD4-4) from Anopheles dirus, Y119E, in Complex with S-Hexyl Glutathione | | Descriptor: | Glutathione transferase GST1-4, S-HEXYLGLUTATHIONE | | Authors: | Wongsantichon, J, Robinson, R.C, Ketterman, A.J. | | Deposit date: | 2009-02-10 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural contributions of delta class glutathione transferase active-site residues to catalysis
Biochem.J., 428, 2010
|
|
3GH6
 
 | |
4S11
 
 | |
4S10
 
 | |
4YJY
 
 | |
5WTS
 
 | |
6IM8
 
 | | CueO-PM2 multicopper oxidase | | Descriptor: | Blue copper oxidase CueO,PM2 peptide,Blue copper oxidase CueO | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
6IM9
 
 | | MDM2 bound CueO-PM2 sensor | | Descriptor: | Blue copper oxidase CueO,PM2 peptide,Blue copper oxidase CueO, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
6IM7
 
 | | CueO-12.1 multicopper oxidase | | Descriptor: | Blue copper oxidase CueO,12.1 peptide,Blue copper oxidase CueO, CALCIUM ION | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
4UMN
 
 | | Structure of a stapled peptide antagonist bound to Nutlin-resistant Mdm2. | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, M06 | | Authors: | Chee, S, Wongsantichon, J, Quah, S, Robinson, R.C, Verma, C, Lane, D.P, Brown, C.J, Ghadessy, F.J. | | Deposit date: | 2014-05-20 | | Release date: | 2014-05-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of a stapled peptide antagonist bound to nutlin-resistant Mdm2.
PLoS ONE, 9, 2014
|
|
5YPU
 
 | | Crystal structure of an actin monomer in complex with the nucleator Cordon-Bleu MET72NLE WH2-motif peptide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Scipion, C.P.M, Wongsantichon, J, Ferrer, F.J, Yuen, T.Y, Robinson, R.C. | | Deposit date: | 2017-11-03 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for the roles of divalent cations in actin polymerization and activation of ATP hydrolysis
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YVN
 
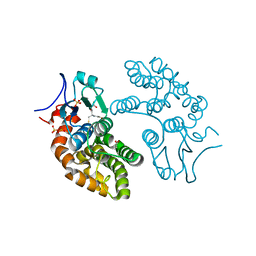 | | Human Glutathione Transferase Omega1 | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase omega-1, ... | | Authors: | Saisawang, C, Ketterman, A, Wongsantichon, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
5YVO
 
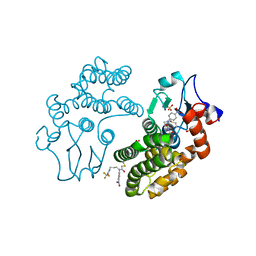 | | Human Glutathione Transferase Omega1 covalently bound to ML175 inhibitor | | Descriptor: | ACETATE ION, Glutathione S-transferase omega-1, N-{3-[(2-chloro-acetyl)-(4-nitro-phenyl)-amino]-propyl}-2,2,2-trifluoro-acetamide, ... | | Authors: | Saisawang, C, Ketterman, A, Wongsantichon, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glutathione transferase Omega 1-1 (GSTO1-1) modulates Akt and MEK1/2 signaling in human neuroblastoma cell SH-SY5Y.
Proteins, 87, 2019
|
|
4P8V
 
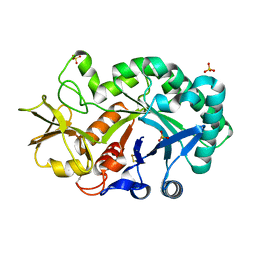 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc2) were solved to resolutions of 1.5 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
4P8X
 
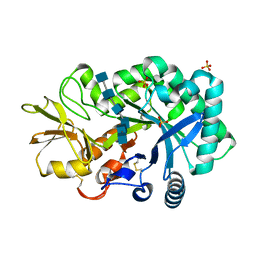 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc6) were solved to resolutions of 2.48 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
4P8U
 
 | | The crystal structures of YKL-39 in the absence of chitooligosaccharides was solved to resolutions of 2.4 angstrom | | Descriptor: | Chitinase-3-like protein 2, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
4P8W
 
 | | The crystal structures of YKL-39 in the presence of chitooligosaccharides (GlcNAc4) were solved to resolutions of 1.9 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 2, SULFATE ION | | Authors: | Suginta, W, Ranok, A, Robinson, R.C, Wongsantichon, J. | | Deposit date: | 2014-04-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Thermodynamic Insights into Chitooligosaccharide Binding to Human Cartilage Chitinase 3-like Protein 2 (CHI3L2 or YKL-39).
J.Biol.Chem., 290, 2015
|
|
1V2A
 
 | | Glutathione S-transferase 1-6 from Anopheles dirus species B | | Descriptor: | GLUTATHIONE SULFONIC ACID, glutathione transferase gst1-6 | | Authors: | Oakley, A.J. | | Deposit date: | 2003-10-10 | | Release date: | 2003-10-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification, characterization and structure of a new Delta class glutathione transferase isoenzyme.
Biochem.J., 388, 2005
|
|
5XXK
 
 | |
